Figure 1.
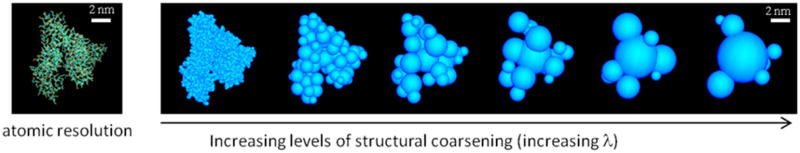
Structural coarse-graining of albumin (based on PDB: 1AO6). Reverse coarsening (fine graining) to any λ level is straightforward (used in Figures 6–8). The hydrodynamic radius RH of albumin in water has been measured at ~3.2–3.48 nm.22,44 For the rigid model used here, n = 7 and ρi = Ri + dw, where dw is the average thickness of the hydration layer surrounding the protein. Using a single layer (dw = 2.8 Å), eq 6 yields RH = 3.33 nm. The calculated translational and rotational diffusion constants are DT ~ 6.6 × 1011 m2 s−1 and DR ~ 4.4 × 106 1 s−1, respectively, close to the experimental values.21 The rotational correlation times τD(1) and τD(2) (Debye’s relaxation time) of the albumin macrodipole (~500 D21) are thus ~33 ns and ~0.1 μs, respectively. The model can be used to calculate dielectric and spectroscopic properties of albumin solutions, although the relaxation times indicate that long dynamics simulations would be needed to properly sample the conformational space, which justifies the choice of Monte Carlo sampling used in this study.
