Figure 7. hnRNPC1/C2 tetramer stability based on species-specific amino acid substitutions.
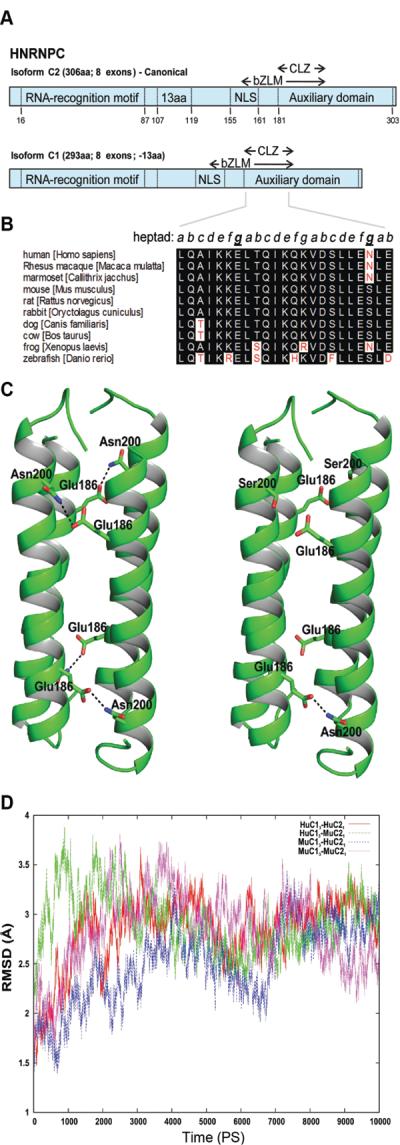
A) Schematic representation of human hnRNPC isoforms. Each protein contains a single RNA recognition motif (RRM), a delineated nuclear localization signal (NLS), a basic leucine zipper-like motif (bZLM), and an encompassing acidic auxiliary domain. hnRNPC2 contains an additional 13 a.a. and is expressed at one-third the level of hnRNPC1. An oligomerization domain (CLZ) is also present in C proteins. Numbers represent a.a. position. B) Alignment of CLZ sequences derived from the Ensembl database (www.ensembl.org). Key asparagine (N) to serine (S) change within the mouse sequences occupying an interhelical contact surface heptad residue within the coiled coil core of hnRNP C. Similar residues are highlighted in black while discrepant residues are highlighted in white. The heptad arrangement is in italics, with the key intermolecular residues forming anti-parallel chain contacts underlined and bolded. Numbering based on the human C2 full-length isoform. C) The hnRNPC protein CLZ tetramers of human (Hu) and mouse (Mu) C1 and C2 proteins. HuC13-HuC21 (left panel) and MuC13-HuC21 (right panel). The Asn200, Ser200 and Glu186 residues are shown in stick representation. In HuC13-HuC21 CLZ domains, pairs from the adjacent helices make four hydrogen bonds in the coiled coil tetramer. In contrast, in the MuC13-HuC21 tetramer, only one hydrogen bond is possible. The hydrogen bonds formed between Asn200 and Glu186 are shown in dashed lines. D) Molecular dynamic simulations for CLZ tetramers. The root mean square fluctuations for the backbone atoms of the tetramer complexes for different combinations of human (Hu) and mouse (Mu) hnRNPC1 and C2. HuC13-HuC21, red; HuC13-MuC21, green; MuC13-HuC21, blue and MuC13-MuC21, magenta. Data are based on experimental structure over 10 nanosec simulations.
