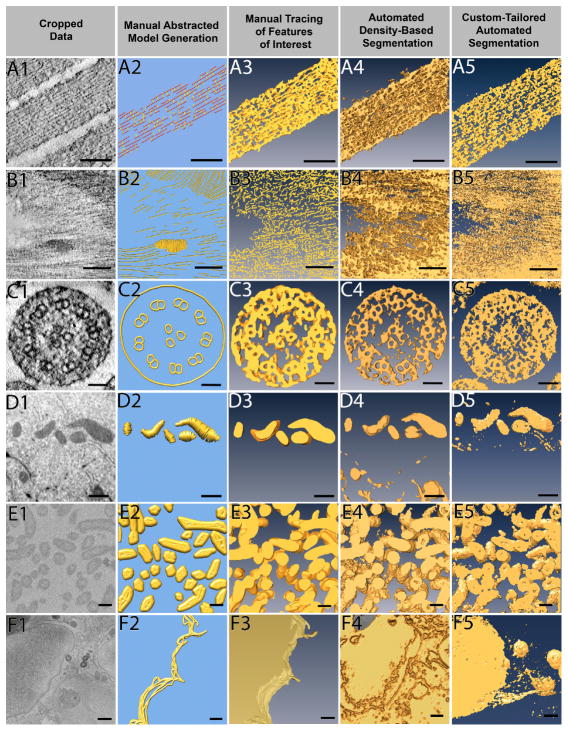
Figure 3. Different data sets being segmented by four different approaches.
Six example data sets were segmented by all four approaches: manual abstracted model generation, manual tracing, automated density-based segmentation, and custom-tailored automated segmentation. Manual abstracted model generation was effective for the resin embedded stained tomography of stereocilia (A), as the purpose was not to extract exact densities but rather to create a model for quantitative purposes. For the resin embedded stained tomography of plant cell wall (B), automated density-based segmentation was the most effective method to quickly extract the cellulose through many slices, where as the manual methods took much more effort on only a few slices of data. Manual abstracted model generation generated the microtubule triplet in the stained tomography of kinocilium (C) while other segmentation methods did not, yet the two automated approaches extracted the densities more quickly and were therefore preferred. Due to the shape of mitochondria from FIB-SEM of breast epithelial cells (D), manual tracing provided the cleanest result, and the low population density combined with use of interpolation methods allowed for quick segmentation. Given the large volume that needed to be segmented, custom-tailored automated segmentation proved to be most efficient to segment the SBF-SEM bacteria data (E), but both automatic approaches were comparable. Although time consuming, the only method to extract the FIB-SEM of breast epithelial cell membrane (F) was manual tracing. Scale Bars: A1–A5 = 100 nm, B1–B5 = 100 nm, C1–C5 = 50 nm, D1–D5 = 500 nm, E1–E5 = 200 nm, F1–F5, bars = 500 nm.