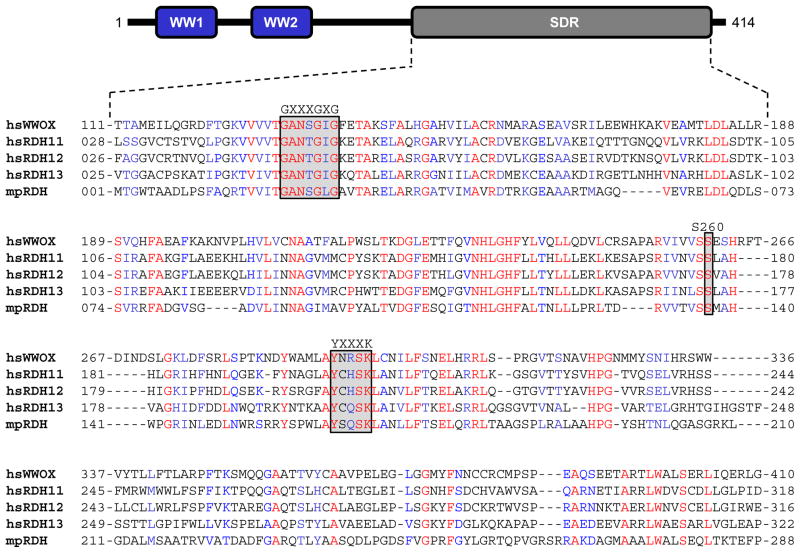
Figure 6.
Amino acid sequence alignment of SDR domains of hsWWOX (Q9NZC7), hsRDH11 (Q8TC12), hsRDH12 (Q96NR8), hsRDH13 (Q8NBN7) and mpRDH (Q741V7) derived from Homo sapiens (hs) and Mycobacterium paratuberculosis (mp). The UniProt ID of each protein is provided in the corresponding parenthesis. The numerals flanking each row of the sequence alignment denote the amino acid position of corresponding protein. Absolutely conserved residues are colored red, highly conserved residues in blue and non-conserved residues in black. The GXXXGXG motif is involved in accommodating the NADP+/NADPH cofactor, while the YXXXK consensus sequence represents the active site motif critical for the enzymatic activity of SDR domains. Additionally, an absolutely conserved serine (S260 in WWOX) is required for the stabilization of retinoid substrate within the active site. Note that the modular architecture of WWOX is overlaid to indicate the relative locations of the N-terminal WW1-WW2 tandem module and the C-terminal SDR domain.