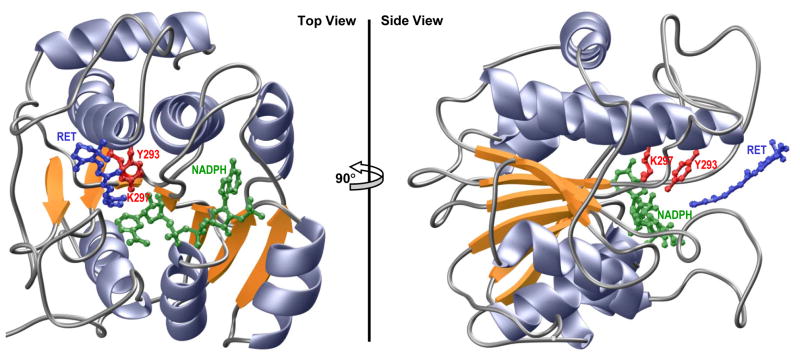
Figure 7.
Ribbon representation of the structural model of SDR domain of WWOX in complex with NADPH cofactor and all-trans-retinal (RET) substrate. Two alternative orientations related by a 90°-rotation about the vertical axis are depicted for the inquisitive eye. In each case, the β-strands are colored yellow, α-helices lavender and the intervening loops gray. The NADPH co-factor and RET substrate are shown in green and blue, respectively. The sidechain moieties of the Y293/K297 catalytic dyad, located within the RXXXK active site motif of WWOX, are depicted in red. Note that the structural model of SDR domain was built in several stages using the MODELLER software based on homology modeling (85). Firstly, the apo-structure of SDR domain was constructed using the crystal structure of mpRDH as a template (PDBID 3RD5). It should be noted here that mpRDH shares greater than 50% sequence similarity with the SDR domain of WWOX, thereby implying that the structural model of the latter can be relied upon with a high degree of confidence. Next, NADPH was mapped to the apo-structure of SDR domain on the basis of its structural homology with the crystal structure of bacterial L-sorbose reductase bound to NADPH (PDBID 3AI2). Finally, RET was docked onto the structure of SDR domain bound to NADPH in analogy with the binding mode of 17β-estradiol to 17β-hydroxysteroid dehydrogenase 1 (PDBID 1FDT) using various distance restraints between RET, NADPH and Y293 proton acceptor/donor located within the YXXXK active site motif. To ensure structural convergence, a total of 100 atomic models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The atomic models were rendered using RIBBONS (86).