Abstract
The effect of electrolyte cation on the unzipping of furan-containing double-stranded DNA in an α-hemolysin (αHL) nanopore is described. The current through an open αHL channel increases in proportion to the ion mobility. However, the ionic current measured during residence of a DNA duplex inside the protein pore shows a more complex dependence on the choice of cation, indicating that the current measured during DNA residence in the pore is modulated by the specific interactions of the cations with the DNA and/or αHL. The residence time (stability) of the DNA duplex inside the pore prior to unzipping is also highly dependent on the cation, in striking contrast to the small variation in duplex stability (as measured by the melting temperature) in bulk electrolyte solution. A missing base in DNA can be detected in the latch region of αHL with optimal current resolution in RbCl, while optimal time resolution is possible in LiCl.
Keywords: nanopore, DNA damage, cations, latch zone, α-hemolysin
The α-hemolysin (αHL) nanopore has emerged as a promising platform for the analysis of DNA 1–9. When single-stranded DNA (ssDNA) is electrophoretically driven through αHL in an electrolytic solution, the current temporarily decreases, relative to the open-channel value, because the DNA blocks the flux of the current-carrying ions. The majority of this resistance is located at a 1.4 nm-diameter central constriction 10 in the middle of the pore (Figure 1A), the dimensions of which are comparable to the ~1 nm diameter of ssDNA.11
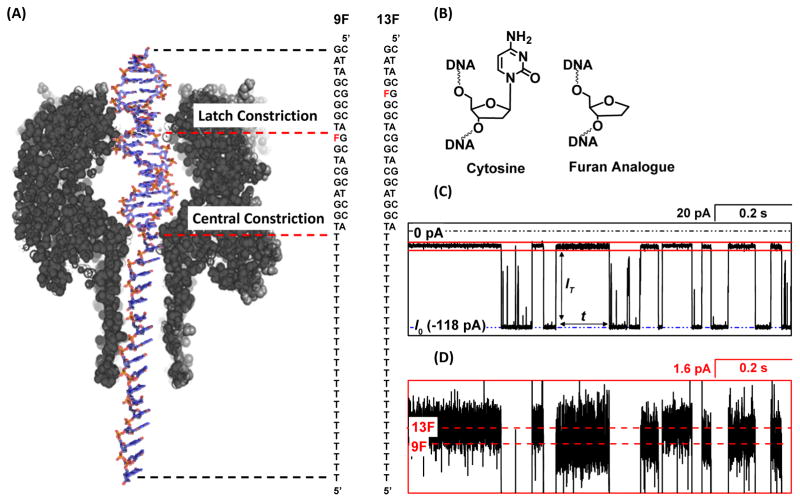
FIGURE 1.
(A) Position of the furan within dsDNA relative to the latch constriction during dsDNA residence for the duplexes 9F and 13F. (B) Replacing the C:G base pair situated at the latch constriction with a furan (abasic site analogue) opposite G increases the ion flux through αHL. (C) Representative I-t trace indicating the change in current from the open channel during dsDNA residence events. (D) Expanded view of the trace outlined in the blue box region in (C) showing the difference in blocking current between duplexes with a furan situated inside (9F) and outside the latch region (13F). Experiments were carried out at 25 °C in a 1.00 M KCl solutions buffered to pH 7.5 using 10 mM phosphate.
Double-stranded DNA (dsDNA) does not translocate freely through the αHL channel because its diameter (2.0 nm) 11 is too large to fit through the central 1.4 nm constriction. However, with appropriate design of the nucleic acid, and at sufficient driving force (>100 mV applied bias), dsDNA will denature (‘unzip’) within the pore releasing the two constituent single-stranded components.12–16 The residence time of the duplex prior to unzipping is dependent on the dsDNA stability, and can be used to identify the presence of base-mismatches,12–14 oxidative damage,15 and abasic sites.17
Ion channel recordings using αHL are generally performed using ~1 M KCl as the supporting electrolyte. Studies with alternative cations are extremely limited for αHL,18–20 and the effects of different cations on the dsDNA residence and unzipping have not previously been studied. Here, we demonstrate that the choice of monovalent cation significantly affects both the dsDNA residence time and the measured blocking current, and thus, the ability to detect the presence of a base modification.
Double-stranded DNA modified with a single-sided ‘threading’ poly-T tail is driven into the αHL channel at the cis opening and up to the central constriction that separates the vestibule and β-barrel (Figure 1A).10 Attenuation of the current through αHL during dsDNA residence (i.e., prior to unzipping) results in ‘blocking events’ that are characterized by a well-defined current level. Previously, we discovered that IT is dependent on the structure of the duplex at the latch constriction, which is situated in the upper vestibule of αHL.17, 21 In these experiments, the sequence of the DNA is either 5′-(T)24TGGAGCTGFTGGCGTAG or 5′-(T)24TGGAGCTGCTGGFGTAG; the complementary strand, 5′-CTACGCCAGCAGCTCCA, is used to form the duplex. A furan group, F, is substituted for a cytosine (Figure 1A and 1B) in the sequence such that it is situated either inside or outside of the latch region, respectively, during dsDNA residence. This particular sequence was chosen because it is a part of the KRAS gene, and unrepaired damage in this sequence have been shown to result in harmful mutations.22
Attenuation of the current from the open channel value, I0, to a blockage current, IT, occurs when dsDNA is driven into the vestibule. We find that IT is 1.6 pA less in 1.00 M KCl electrolyte when a furan is situated in the latch constriction relative to a G:C base-pair at 25 °C (Figure 1C and 1D). This current difference is the basis for detection of a missing base, and has been applied previously to monitor the kinetics of the repair enzyme uracil-DNA glycosylase (UDG), which removes the abnormal component uracil from DNA to leave an abasic site.17
The finding that the blockage current for dsDNA is very sensitive to the presence or absence of a base at the latch constriction is surprising, as it has been previously reported that the majority of the resistance to the ion flux is located at the central constriction.23 We decided to first investigate how the total resistance is distributed between the latch and central constriction when the αHL vestibule is occupied by dsDNA with a tail that extends through the central constriction, as shown in Figure 1A.
In a typical experiment, 15 μM of the duplexes 9F and 13F, and 15 μM excess of 13F 41-mer (present as ssDNA) were added to the cis side of αHL and a bias of 120 mV was applied to stochastically capture both individual duplexes and ssDNA. The capture and unzipping events of the 9F and 13F duplexes, along with ssDNA translocation events (from the excess 13F 41-mer) were recorded. The type of event (either ssDNA translocation or dsDNA unzipping) can easily be differentiated based on the event time, τ. Translocation of the ssDNA 41-mer in 1.00 M KCl falls in the range of 80 to 820 μs, while residence of dsDNA prior to unzipping is ≥ 5 ms.15–17
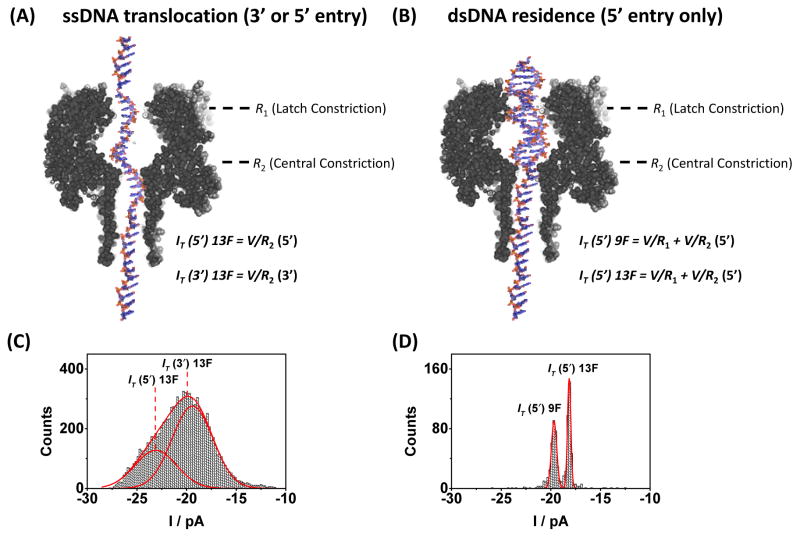
In a simplistic model, the current measured while the dsDNA resides in the pore prior to unzipping can be attributed to the ionic resistances at two sites within the αHL nanopore: (1) the latch constriction (R1), which is specific to dsDNA, and (2) the 1.4 nm central constriction (R2) 10. We assume that R1 is negligible for ssDNA, as the constriction size (2.6 nm) 10 is significantly larger than the diameter of ssDNA (1 nm).11
Consider first ssDNA translocation (Figure 2A), where the current is dominated by R2 (RT ~ R2). Two peaks are observed in the current histogram (Figure 2C) because ssDNA can enter from either the 3′ of 5′ direction. It has been previously established that entry from the 5′ end results in less attenuation of the ion flux than entry from the 3′ end.16, 24 The resistance at the central constriction for ssDNA translocation was calculated from the Gaussian peaks in Figure 2C to be 5.2 ± 0.2 GΩ for 5′ entry, and 6.3 ± 0.2 GΩ for 3′ entry.
FIGURE 2.
Relative contribution to the ion channel current during ssDNA translocation and dsDNA residence in 10 mM phosphate buffer (pH 7.5), 1.00 M KCl electrolyte at 25°C. (A) Structure of the αHL channel overlaid with a 41-mer heteropolymer. (B) Structure of the αHL channel and dsDNA prior to unzipping. Entry is by the 5′ end of the tail only. (C) The measured current (IT) for ssDNA translocation is largely determined by the resistance at the central constriction (R2) and is dependent on the direction of ssDNA entry. (D) For dsDNA residence, IT is a function of the resistance at both the latch constriction (R1) and the central constriction (R2). Moving the furan into the latch during unzipping (9F) reduces R1 and increases IT. Counts indicate either (C) the number of ssDNA translocation events or (D) the number of dsDNA unzipping events.
For dsDNA residence (Figure 2B), the current is dependent on both R1 and R2 (RT = R1 + R2). The total measured current during dsDNA residence is shown in Figure 2D, and is dependent on the structure of DNA at the latch constriction site (i.e., if a furan is present or absent). The resistance at the latch constriction during occupation by duplexes 9F and 13F can be estimated by subtracting the total resistance across αHL from the known resistance at the central constriction (entry of the dsDNA into αHL occurs only from the 5′ end of the tail). We find that R1 = 1.4 ± 0.2 GΩ when a G:C base-pair (duplex 13F) is located in the latch constriction, and that this value decreases to 0.9 ± 0.4 GΩ when the latch is occupied by a G opposite to a F (duplex 9F). Thus, the fractional percentages of the total resistance corresponding to the central and latch constrictions for duplex 13F are 79 ± 3% and 21 ± 3%, respectively. For duplex 9F, the fraction percentage to the total resistance at the central and latch constrictions are 86 ± 3% and 14 ± 3%, respectively. The significant fraction of the total resistance located at the latch constriction, when occupied by dsDNA, allows base modifications in this location to attenuate the current to different degrees that are readily measured.
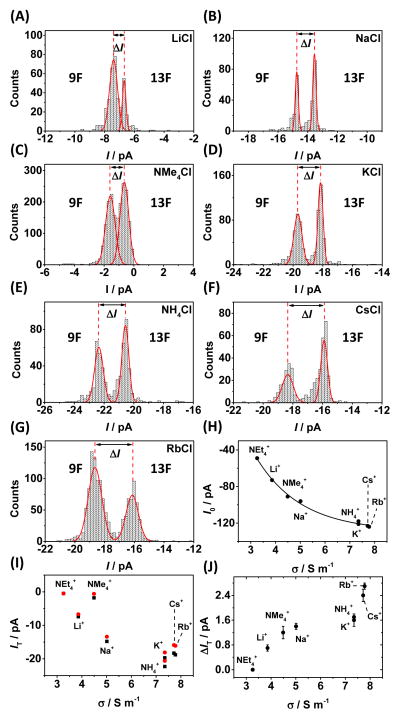
We performed a series of ion channel recordings in eight different electrolyte solutions, all at a concentration of 1.00 M, in which the anion (Cl−) was kept constant while the cation was varied. Figure 3 shows the current through the open αHL channel (I0) as a function of conductivity (σ). Ions with a lower conductivity (e.g., Li+) are typically strongly hydrated, and their mobility is therefore reduced. This gives rise to the lower open channel currents measured in these electrolytes.
FIGURE 3.
The effect of cation on discrimination between duplexes 9F and 13F. (A–G) current histograms showing the measured blocking current (IT) and current difference (ΔIT) for the cations studied. (H) The effect of conductivity on the measured current through an open αHL protein channel, I0, and (I) during dsDNA residence, IT for duplexes 9F (black squares) and 13F (red circles). (J) The dependence of ion conductivity on the current difference for duplexes 9F and 13F on electrolyte conductivity. Experiments were carried out in 10 mM phosphate buffer (pH 7.5) at 25 °C with electrolyte added at 1.00 M concentration as indicated. Counts indicate the number of dsDNA unzipping events.
The situation when dsDNA occupies the pore is more complex. Overall, the measured current during DNA residence decreases as cation mobility decreases, following the same trend as for the open channel current. However, the current measured using Cs+, tetramethlyammonium (Me4N+) (and to a lesser extent, Rb+) do not follow the general trend. The measured current through the pore during DNA residence for these ions is less than would be expected based on the trend in the open channel current as a function of cation. Cs+ and Rb+ have mobilities similar to K+, and the hydrated radii of these ions are also similar, with values of 0.331, 0.329, and 0.329 nm reported for K+, Rb+, and Cs+, respectively.25 Assuming the diameter of ssDNA is approximately 1 nm (half that of B-form dsDNA),11 and given that the diameter of the central constriction is 1.4 nm,10 then the remaining open space at this point is ~0.4 nm, which is smaller than the hydrated diameter of all the ions studied. The central constriction is formed of glutamate and lysine residues 26, both of which are charged at pH 7.5, as is the DNA itself. Strong interaction of cations with these significant regions of charge seems highly plausible, and in such a scenario, the ions may undergo at least partial dehydration and/or rearrangement of their solvent shell in order to transit through the pore. Thus, the ionic radii of the ions need to be considered. The ionic radii of K+, Rb+, and Cs+ are 0.133, 0.148, and 0.169 nm, respectively.27 We speculate that a size-exclusion effect is observed for Rb+ and Cs+ ions, which reduces the conductivity (increases resistance) at the central constriction. The same argument also applies to the Me4N+ ion, which is comparable in size (0.285 nm radius 25) to the free space at the central constriction during DNA residence (0.4 nm). For the Et4N+ ion, which has an ionic radius of 0.348 nm,25 near-zero current is observed during both dsDNA residence and ssDNA translocation (Figure S1), suggesting that Et4N+ is too large to pass between the DNA and the protein interior surface.
The measured current difference (ΔIT) for the resident DNA with a furan situated at the latch region (9F) relative to the F placed outside the latch region (13F) is shown in Figure 3A–G and summarized in Figure 3I and J. Weakly hydrated, more mobile ions give the largest current difference. In a mechanistic sense, this indicates the more mobile ions are able to exploit the additional pathway created by the absence of the base at the latch constriction most effectively. The largest current difference is observed for Rb+, indicating that this cation is the optimal choice for identifying the presence of a furan at the latch constriction based on ion current signatures.
While the electrolyte cation does effect the measured current during dsDNA residence, no effect is observed for electrolytes containing the NO3− anion instead of Cl− (Figure S2), because anions are largely excluded from the pore when DNA is present.24, 28–31 This topic is discussed in more detail in the supplementary material.
The residence time of dsDNA in the protein pore prior to unzipping is also strongly dependent on the choice of cation (Figure 4), despite the very limited changes in dsDNA stability (Tm changes by less than 2 °C across the Group 1 metal ions) for these duplexes in bulk solution (Table S1). The residence time inside the αHL channel decreases with increasing ion mobility; low mobility, strongly hydrated ions (e.g., Li+ and Na+) significantly stabilize DNA residing in the αHL channel. The largest resolution in unzipping times between duplexes 9F and 13F is observed for LiCl (Δτ = 420 ± 80 ms). While the residence time of dsDNA inside the duplex is cation dependent, the capture rate is not, with an average of 6 ms between capture events for all the salts studied when the dsDNA concentration was 15 μM. To date, all studies reporting dsDNA unzipping times (τ) have been performed in KCl solutions, conditions that yield much smaller differences in unzipping times between two duplexes of similar stability. These results make a clear case for the use of LiCl for identifying dsDNA if the identification is based on residence time.
FIGURE 4.
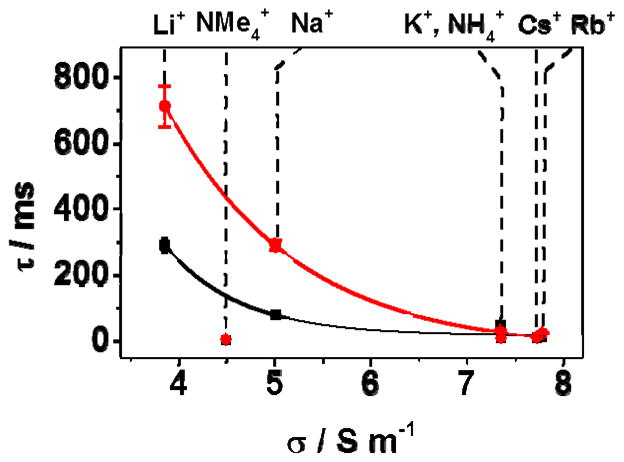
The effect of cation conductivity on the measured duplex unzipping time when a furan site is present at the latch (duplex 9F, black squares) and outside the latch (duplex 13F, red circles). The cation type is indicated on the upper y-axis. Tm measurements for each salt are given in Table S1. Experiments were carried out in 10 mM phosphate buffer (pH 7.5) at 25 °C with electrolyte added at 1.00 M concentration as indicated. Data were recorded at 120 mV (trans vs. cis). Time distribution histograms for each cation are shown in Figure S3.
The order of unzipping times (τ) as a function of cation follows the series below, with unzipping times for the duplex 13F decreasing from 714 ± 62 ms in LiCl to 6 ± 1 ms in Me4N+:
Previous studies have shown that at high electrolyte concentration (> 1 M), anions have a larger effect on the stability of dsDNA than cations.32–33 However, in the anion-free environment of the αHL pore during dsDNA residence, the cation specific interactions with DNA secondary structure can be expected to be more significant than in bulk solution. Several studies 34–36 have shown that the strength of monovalent counterion binding to DNA is specific to the type of cation, and this will affect the ability of the ions to screen neighboring phosphate charges within a dsDNA molecule and increase duplex stability. Stronger counterion binding to DNA may also reduce the effective driving force on DNA as it is transported through the pore.37 Excluded from the preceding discussion is tetraethylammonium cation, Et4N+. In the presence of this large hydrophobic cation, unzipping of dsDNA in αHL is either not possible or very slow (near-complete blockage of the channel was observed for up to 2 minutes), suggesting the effective force on dsDNA inside the αHL pore in the presence of Et4N+ is small. Despite this, translocation of ssDNA still occurs in the Et4NCl electrolyte, although the current is essentially completely blocked. (Figure S1). Et4N+, which is ~0.4 nm in radius, likely does not fit through the constriction of αHL when occupied by ssDNA.
In conclusion, we have demonstrated that the choice of electrolyte cation has a significant effect on the measured current through αHL when dsDNA resides within the vestibule. Changes in the measured current are attributed primarily to the intrinsic conductivity of the cation, but specific ion-DNA and/or ion-protein interactions have a noticeable effect for ions of similar conductivity (K+, Rb+, Cs+). Optimal detection of a furan site in dsDNA at the latch constriction of αHL, relative to a fully-complementary reference is achieved in RbCl, a discovery that will have a significant impact on the ability to sense structural changes in dsDNA using the latch constriction of αHL. The residence time of dsDNA in the pore prior to unzipping is also strongly dependent on the cation with smaller, mobile cations significantly stabilizing the dsDNA within the pore and increasing residence time prior to unzipping.
Supplementary Material
Acknowledgments
RPJ acknowledges funding from a Marie Curie International Outgoing Fellowship under the EU FP7 programme (Project No. 625984). The work was funded by a grant from the National Institutes of Health (GM093099). The authors thank Electronic BioSciences Inc. (San Diego, CA) for donating the ion-channel recording instrument and software.
Footnotes
Effect of electrolyte anion on detecting damage with the latch region of αHL. Unzipping time histograms, and tabulated data. Translocation of dC60 ssDNA homopolymer in a Et4N+ electrolyte. Full experimental details. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond Time-Scale Discrimination among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments within Single RNA Molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clamer M, Höfler L, Mikhailova E, Viero G, Bayley H. Detection of 3′-End Rna Uridylation with a Protein Nanopore. ACS Nano. 2013;8:1364–1374. doi: 10.1021/nn4050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasianowicz JJ, Robertson JWF, Chan ER, Reiner JE, Stanford VM. Nanoscopic Porous Sensors. Ann Rev Anal Chem. 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 5.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc Natl Acad Sci USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purnell RF, Schmidt JJ. Discrimination of Single Base Substitutions in a DNA Strand Immobilized in a Biological Nanopore. ACS Nano. 2009;3:2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 7.Wanunu M. Nanopores: A Journey Towards DNA Sequencing. Phys Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Rapid Discrimination among Individual DNA Hairpin Molecules at Single-Nucleotide Resolution Using an Ion Channel. Nat Biotech. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 9.Vercoutere WA, Winters-Hilt S, DeGuzman VS, Deamer D, Ridino SE, Rodgers JT, Olsen HE, Marziali A, Akeson M. Discrimination among Individual Watson–Crick Base Pairs at the Termini of Single DNA Hairpin Molecules. Nuc Acids Res. 2003;31:1311–1318. doi: 10.1093/nar/gkg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 11.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA Dodecamer: Conformation and Dynamics. Proc Natl Acad Sci USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping Kinetics of Double-Stranded DNA in a Nanopore. Phys Rev Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 13.Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore Unzipping of Individual DNA Hairpin Molecules. Biophys J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland TC, Dinsmore MJ, Kraatz HB, Lee JS. An Analysis of Mismatched Duplex DNA Unzipping through a Bacterial Nanopore. Biochem Cell Biol. 2004;82:407–412. doi: 10.1139/o04-005. [DOI] [PubMed] [Google Scholar]
- 15.Jin Q, Fleming AM, Ding Y, Burrows CJ, White HS. Structural Destabilization of DNA Duplexes Containing Single-Base Lesions Investigated by Nanopore Measurements. Biochem. 2013;52:7870–7877. doi: 10.1021/bi4009825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Q, Fleming AM, Burrows CJ, White HS. Unzipping Kinetics of Duplex DNA Containing Oxidized Lesions in an α-Hemolysin Nanopore. J Am Chem Soc. 2012;134:11006–11011. doi: 10.1021/ja304169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Q, Fleming AM, Johnson RP, Ding Y, Burrows CJ, White HS. Base-Excision Repair Activity of Uracil-DNA Glycosylase Monitored Using the Latch Zone of α-Hemolysin. J Am Chem Soc. 2013;135:19347–19353. doi: 10.1021/ja410615d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya S, Muzard J, Payet L, Mathé J, Bockelmann U, Aksimentiev A, Viasnoff V. Rectification of the Current in α-Hemolysin Pore Depends on the Cation Type: The Alkali Series Probed by Molecular Dynamics Simulations and Experiments. J Phys Chem C. 2011;115:4255–4264. doi: 10.1021/jp111441p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shim JW, Tan Q, Gu LQ. Single-Molecule Detection of Folding and Unfolding of the G-Quadruplex Aptamer in a Nanopore Nanocavity. Nuc Acids Res. 2009;37:972–982. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An N, Fleming AM, White HS, Burrows CJ. Crown Ether–Electrolyte Interactions Permit Nanopore Detection of Individual DNA Abasic Sites in Single Molecules. Proc Natl Acad Sci USA. 2012;109:11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson Robert P, Fleming Aaron M, Jin Q, Burrows Cynthia J, White Henry S. Temperature Electrolyte Optimization of the α-Hemolysin Latch Sensing Zone for Detection of Base Modification in Double-Stranded DNA. Biophys J. 2014;107:924–931. doi: 10.1016/j.bpj.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeifer G, Besaratinia A. Mutational Spectra of Human Cancer. Hum Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howorka S, Bayley H. Probing Distance and Electrical Potential within a Protein Pore with Tethered DNA. Biophys J. 2002;83:3202–3210. doi: 10.1016/S0006-3495(02)75322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathé J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation Discrimination of Single-Stranded DNA inside the α-Hemolysin Membrane Channel. Proc Natl Acad Sci USA. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkov AG, Paula S, Deamer DW. Two Mechanisms of Permeation of Small Neutral Molecules and Hydrated Ions across Phospholipid Bilayers. Bioelectrochem Bioenerg. 1997;42:153–160. [Google Scholar]
- 26.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced Translocation of Single DNA Molecules through α-Hemolysin Nanopores by Manipulation of Internal Charge. Proc Natl Acad Sci USA. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson RA, Stokes RH. Electrolyte Solutions. Dover Publications Inc; Mineola, New York: 2002. (Second Revised Edition) [Google Scholar]
- 28.Bonthuis DJ, Zhang J, Hornblower B, Mathé J, Shklovskii BI, Meller A. Self-Energy-Limited Ion Transport in Subnanometer Channels. Phys Rev Lett. 2006;97:128104. doi: 10.1103/PhysRevLett.97.128104. [DOI] [PubMed] [Google Scholar]
- 29.Markosyan S, De Biase PM, Czapla L, Samoylova O, Singh G, Cuervo J, Tieleman DP, Noskov SY. Effect of Confinement on DNA, Solvent and Counterion Dynamics in a Model Biological Nanopore. Nanoscale. 2014;6:9006–9016. doi: 10.1039/c3nr06559f. [DOI] [PubMed] [Google Scholar]
- 30.De Biase PM, Solano CJF, Markosyan S, Czapla L, Noskov SY. Bromoc-D: Brownian Dynamics/Monte-Carlo Program Suite to Study Ion and DNA Permeation in Nanopores. J Chem Theor Comp. 2012;8:2540–2551. doi: 10.1021/ct3004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guy Andrew T, Piggot Thomas J, Khalid S. Single-Stranded DNA within Nanopores: Conformational Dynamics and Implications for Sequencing; a Molecular Dynamics Simulation Study. Biophys J. 2012;103:1028–1036. doi: 10.1016/j.bpj.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamaguchi K, Geiduschek EP. The Effect of Electrolytes on the Stability of the Deoxyribonucleate Helix. J Am Chem Soc. 1962;84:1329–1338. [Google Scholar]
- 33.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen PE, Nordén B, Gräslund A. Ionic Effects on the Stability and Conformation of Peptide Nucleic Acid Complexes. J Am Chem Soc. 1996;118:5544–5552. [Google Scholar]
- 34.Bleam ML, Anderson CF, Record MT. Relative Binding Affinities of Monovalent Cations for Double-Stranded DNA. Proc Natl Acad Sci USA. 1980;77:3085–3089. doi: 10.1073/pnas.77.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong A, Yan Z, Huang Y, Wu G. A Solid-State 23Na NMR Study of Monovalent Cation Binding to Double-Stranded DNA at Low Relative Humidity. Mag Res Chem. 2008;46:308–315. doi: 10.1002/mrc.2136. [DOI] [PubMed] [Google Scholar]
- 36.Denisov VP, Halle B. Sequence-Specific Binding of Counterions to B-DNA. Proc Natl Acad Sci USA. 2000;97:629–633. doi: 10.1073/pnas.97.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing Down DNA Translocation through a Nanopore in Lithium Chloride. Nano Lett. 2012;12:1038–1044. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.