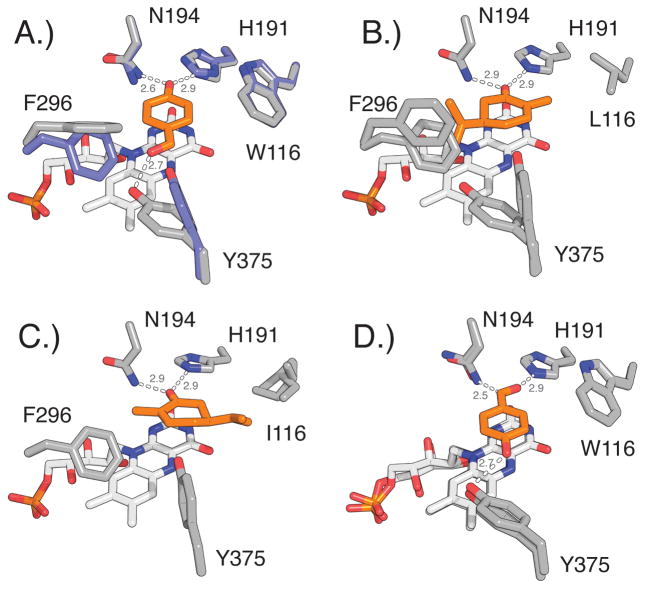
Figure 4.
Conformational changes at positions 116, 296, and 375 upon substrate binding in native OYE1 and OYE variants. Ligands are highlighted in orange A.) Overlay of OYE1 (blue, PDB: 1OYA) and OYE1 with bound HBA (gray, PDB: 1K03) shows the reorientation of F296 (90° rotation of phenyl ring) and Y375 (side chain rotation). B.) OYE1(W116L) with bound (R)-carvone (PDB: 4GWE) as model for carvone binding in normal orientation. C.) OYE1(W116I) with bound (S)-carvone (PDB: 4GE8) shows substrate in flipped orientation. The re-positioning of the isopropenyl group near I116 eliminates the need for conformational changes of F296 and Y375. D.) Overlay of cpOYE303 holoenzyme (PDB: 4RNU) and cpOYE303 with bound HBA (PDB: 4RNV). Hydrogen bonding interactions and distances (in angstrom) are indicated.