Figure 5. iPOP for personalized medicine.
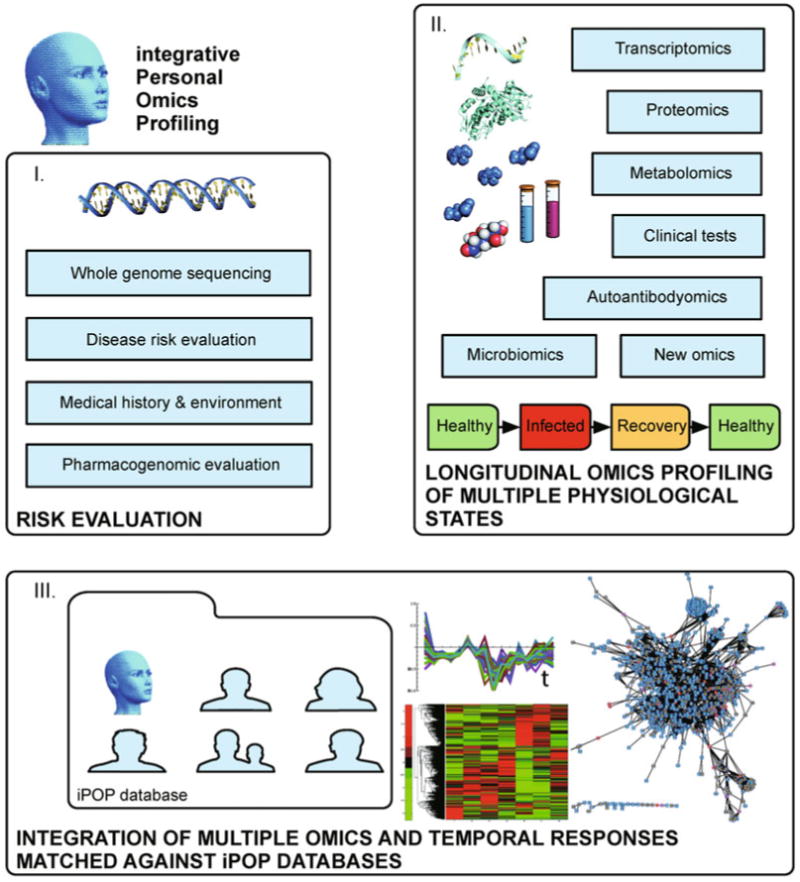
The framework described in the text employs multi-omics analyses (see above and Figures 1–4) that may be implemented for individuals. In step I) Risk estimation for disease is carried out using a whole genome sequencing to perform variant analysis coupled to medical history, environmental considerations and pharmacogenomics evaluations. In step II) Dynamic profiling of multiple omics using an array of technologies follows multiple omics longitudinally in a subject as they progress through their different physiological states, including healthy, disease, and recovery states. Thus thousands of molecular components are collected over time for III) Data integration and biological impact assessment, using temporal patterns to obtain matched omics information, correlate and classify responses, compare against pathway databases and visualize components, e.g., current pathway tools include DAVID [206,272], KEGG [151], Reactome [157–161], Ingenuity Pathway Analysis (IPA); networks can be visualized using Cytoscape [207], various R packages through Bioconductor [234], Matlab by MathWorks and several others. The future iPOP implementations may be gathered into a curated database of iPOP-disease associations that may help in categorizing an omics dynamic response to a catalogued physiological state and disease onset, with potential diagnostic capabilities.
