Abstract
Schizophrenia (SZ) is a devastating mental disorder afflicting 1% of the population. Recent genome-wide association studies (GWASs) of SZ have identified >100 risk loci. However, the causal variants/genes and the causal mechanisms remain largely unknown, which hinders the translation of GWAS findings into disease biology and drug targets. Most risk variants are noncoding, thus likely regulate gene expression. A major mechanism of transcriptional regulation is chromatin remodeling, and open chromatin is a versatile predictor of regulatory sequences. MicroRNA-mediated post-transcriptional regulation plays an important role in SZ pathogenesis. Neurons differentiated from patient-specific induced pluripotent stem cells (iPSCs) provide an experimental model to characterize the genetic perturbation of regulatory variants that are often specific to cell type and/or developmental stage. The emerging genome-editing technology enables the creation of isogenic iPSCs and neurons to efficiently characterize the effects of SZ-associated regulatory variants on SZ-relevant molecular and cellular phenotypes involving dopaminergic, glutamatergic, and GABAergic neurotransmissions. SZ GWAS findings equipped with the emerging functional genomics approaches provide an unprecedented opportunity for understanding new disease biology and identifying novel drug targets.
Keywords: schizophrenia, genomics, open chromatin, microRNA, iPSC, neurons, genome editing
Introduction
Although schizophrenia (SZ) symptoms can be improved by current medications, there is a need for more effective treatments. Most available antipsychotic drugs are still based on the blockade of dopamine D2 receptors (DRD2s), a mechanism discovered over 50 years ago[1]. Recent SZ genome-wide association studies (GWASs) have identified >100 significant genome-wide susceptibility loci with common variants associated with disease[2–7], providing an unprecedented opportunity to understand new disease biology and identify novel drug targets. The genome-wide approach has also implicated multiple rare and large recurrent copy number variations (CNVs) of larger effect size in an increasing risk for developing SZ[8–10].
Although large-scale exome sequencing in SZ has not identified specific rare/low-frequency genetic variants or genes associated with SZ[11, 12], these studies still revealed biological insights consistent with SZ GWAS and CNV studies. This review summarizes the leading biological insights from these genetic findings and discusses conceptual and technical challenges and opportunities in understanding the disease biology underlying the exciting genetic discoveries.
Success of SZ GWAS
In the past five years, we have witnessed the success of SZ GWAS[2–7], an unbiased approach to interrogate the entire genome for SZ risk loci. SZ GWASs have demonstrated the polygenic nature of SZ, each gene contributing a small to moderate effect[3]. With 36,989 SZ cases and 113,075 controls[7], SZ GWAS yielded unparalleled increases of independent genome-wide significant risk loci, from the previously reported 7 to the current 108 independent SZ risk loci. These discoveries not only establish the significant genome-wide association with the DRD2 locus[7], which is central to the classical dopaminergic hypothesis of SZ pathogenesis, but also identify the enrichment of associations with genes involved in neuronal calcium signaling, dendritic spines, and post-synaptic densities[6, 7], highlighting the importance of glutamatergic neurotransmission. Most loci represent new disease biology.
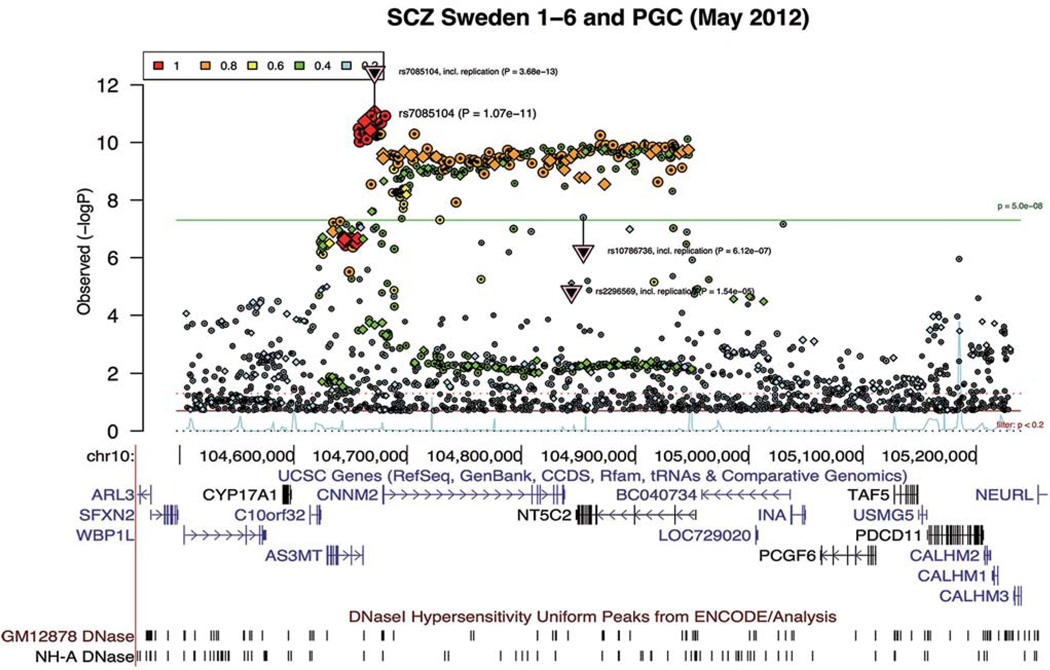
Albeit the success of SZ GWAS, challenges remain. Besides the “missing” heritability, a substantial proportion of genetic risk remains unexplained; one major challenge is to understand the causal molecular mechanisms underlying these associations. This has been hampered by the fact that each risk locus often spans multiple genes and contains many equally-associated single-nucleotide polymorphisms (SNPs) (i.e., due to linkage disequilibrium, LD; r2 >0.8) with SZ (Fig. 1). This makes it difficult to determine the causal variant and what gene(s) is affected. For instance, the GWAS association at the DRD2 locus[7] spans not only DRD2, but also the adjacent NCAM1, a gene important for neurite outgrowth. Thus, whether SZ GWAS at this locus supports the classical dopaminergic hypothesis remains uncertain. Similarly, the SZ risk locus at chr10[6] (Fig. 1) contains hundreds of equally-associated SNPs, and spans >10 genes of which three have synaptic functions: INA, CALHM1, and NEURL. It thus remains to be tested whether the causal variants at this locus influence genes with synaptic functions. Therefore, despite the exciting GWAS findings, there is a need to identify which of the GWAS-implicated variants are functional and causal, as well as which genes are affected and how.
Fig. 1.
Schizophrenia GWAS locus at chr10 spans hundreds of significant genome-wide SNPs in strong linkage disequilibrium (LD) and multiple candidate genes. Regional association plot and the LD information (color-coded r2) were downloaded from Ricopili (http://www.broadinstitute.org/mpg/ricopili/)[6]. UCSC genome browser (hg19) gene track and open chromatin (DNaseI hypersensitive sites) tracks are shown below the association plot.
SZ GWASs Implicate Abnormal Synaptic Plasticity and Glutamatergic Neurotransmission
The exact pathophysiology of SZ remains unclear. Although multiple major neurotransmitter systems (dopaminergic, glutamatergic, and GABAergic) may be involved, SZ GWASs suggest a major role for glutamatergic neurotransmission, neuronal calcium signaling, and morphological changes (dendritic spines and post-synaptic densities)[6, 7]. This is not to say that other neurotransmitter systems are not important for SZ pathogenesis; for instance, one of the genome-wide significant SZ GWAS loci spans DRD2, a gene central to the classical dopaminergic hypothesis of SZ[1]. However, compared with other neurotransmitter systems, many more GWAS-implicated genes are involved in glutamate neurotransmission. Out of the 108 SZ risk loci, eight contain genes related to synapses or excitatory neurotransmission (Table 1). Indeed, multiple lines of evidence suggest that SZ is a neurodevelopmental disorder with impaired frontal cortical development[11–16]. Whole-exome sequencing[11, 12] and CNV studies[10] also support the role of abnormal synaptic plasticity and glutamatergic neurotransmission in SZ. Exome sequencing in 2 536 SZ cases and 2 543 controls demonstrated that rare disruptive mutations are enriched in gene sets associated with the voltage-gated calcium channel and the signaling complex formed by the activity-regulated cytoskeleton-associated scaffold protein of the postsynaptic density[11]. Another large-scale exome sequencing of SZ trios has shown that de novo mutations are over-represented in glutamatergic postsynaptic proteins comprising activity-regulated cytoskeleton-associated protein and N-methyl-d-aspartate receptor complexes[12]. In terms of CNVs, there is also an increased burden of the largest CNVs (>500 kb) in genes present in the postsynaptic density[10].
Table 1.
SZ GWAS genes associated with synapses
| Chr | P-value | Gene | Synaptic function |
|---|---|---|---|
| 4 | 3.441E-08 | CLCN3 | Chloride channel; synaptic plasticity |
| 5 | 1.305E-09 | GRIA1 | AMPARs; synaptic plasticity |
| 7 | 2.358E-14 | ELFN1 | Postsynaptic protein |
| 10 | 5.523E-17 | CALHM1 | Calcium channel; neural excitability |
| INA | Transport to axons and dendrites | ||
| NEURL | Neurogenesis | ||
| 12 | 1.298E-17 | CACNA1C | Calcium channel; neurotransmission |
| 16 | 1.075E-09 | GRIN2A | Mediator of synaptic plasticity |
| 17 | 1.04E-09 | SRR | Activator of NMDARs |
| 22 | 8.076E-12 | CACNA1I | Calcium channel; synaptic plasticity |
These genetic findings converge with previous pathophysiological evidence of abnormalities of synaptic neurotransmission in SZ. SZ patients show reduced cortical grey matter volume and thickness, as well as reduced functional cortical connectivity[17–20]. Reductions in dendritic spine density are thought to directly contribute to these abnormalities[17, 18, 21, 22]. Specifically, reduced spine density on cortical pyramidal neurons has been reported in SZ[15, 22, 23], and cognitive function in humans has been intimately linked to dendritic spine morphology and density[15, 22, 23]. Dendritic spines, mushroom-shaped protrusions, are the sites of most of the excitatory synapses on pyramidal neurons in the mammalian forebrain[24, 25]. Spine plasticity contributes to the neural circuit remodeling that is crucial for postnatal cognitive development[26, 27]. Altered synaptic plasticity and abnormal synaptic neurotransmission provide a basis for prioritizing synaptic genes for mechanistic studies of SZ biology. The biological insights from GWASs and other SZ genetics findings further inform the cellular phenotypes to characterize in disease modelling.
Gene Regulation as A Causal Molecular Mechanism Underlying the SZ Genetic Findings
Variations in expression are expected to be as influential as changes in protein structure in shaping human-specific brain functions[28, 29]. In SZ, the best case for the importance of gene expression regulation is the gene dosage effect of SZ-associated rare CNVs of high penetrance[8–10]. For most CNVs, although it remains uncertain which gene deletion or duplication is the “driver” of the SZ disease phenotype, it is clear that a 2-fold expression difference as a result of heterozygous deletion or a 1.5-fold expression difference as a result of heterozygous duplication can produce pronounced disease phenotypes. Recent SZ GWAS and exome sequencing further highlight the pivotal role of gene regulation in the causal mechanisms of SZ. Most risk variants are noncoding, and only ~10% of the >100 SZ GWAS risk loci have associations possibly explained by protein-coding SNPs[7], implying that most SZ causal variants may influence the expression of nearby (cis) genes. Exome sequencing of large SZ samples also suggests a limited role of rare coding variants in disease etiology[11, 12], further strengthening the importance of rare noncoding variants.
Transcriptional Regulation
Gene expression is regulated at the transcriptional and post-transcriptional (RNA decay and protein synthesis) levels, so noncoding variants can influence gene expression through both transcriptional and post-transcriptional regulatory mechanisms[30–32]. Compared to RNA decay, transcription remains the predominant mechanism determining individual expression variation[31]. Expression quantitative trait locus (eQTL) mapping can identify variants associated with gene expression[33, 34]. SZ risk loci (under a polygenic model and using SNPs with P <0.5) are enriched for cis-eQTL[35], thus likely conferring disease risk through influencing transcript abundance. However, cis-eQTL mapping in a sizable postmortem brain sample only identified eQTLs that could explain two genome-wide significant SZ loci[7]. The eQTL study may be improved by a larger brain sample, but it will still be limited by the well-known confounding factors associated with using postmortem brain tissue[36] and suboptimal cell types or developmental stages[37–39]. After all, eQTL analysis is still an association-based indirect test rather than directly pointing to specific functional variants.
It has been a challenge to interpret the functional noncoding sequences and predict specific regulatory variants. Classical comparative genomics predicts the regulatory function of a sequence based on its evolutionary conservation, however, sequence conservation and function are often discordant[40]. The recent ENCODE Project and Roadmap Epigenomics Program[41–43] provide rich empirical resources of chromatin state marks and transcription factor binding sites (TFBSs) in 349 cell and tissue samples for the bioinformatic annotation of functional noncoding sequences[44]. These genome-wide chromatin marks can help to predict promoters, enhancers, insulators, and TFBSs. One of the most commonly used chromatin marks is DNaseI hypersensitive sites (DHSs) of chromatin, also called accessible or open chromatin[44]. Mammalian DNAs are tightly coiled and compacted in the form of chromatin, which is a characteristic structure of repeating units of nucleosomes, each with ~200 bp of DNA winding around histone proteins[45]. The extent of chromatin compaction affects the ability of transcription factors and other protein regulators to access the regulatory sequence. Accessible or open chromatin is associated with active transcription. The chromatin state is a dynamic process with multilevel control[46], in which transcription factor binding has been suggested to be the primary driving force. Open chromatin is also correlated with epigenomic histone modifications associated with active enhancers and promoters (e.g., H3K4me1 and H3K4me3)[47–51]. In addition, >60% of methylation-eQTLs[52] are within open chromatin. A major determinant of transcription is chromatin accessibility[41, 51], and open chromatin overlies >97% of cis-regulatory sequences[41, 51, 52]. Open chromatin is thus a versatile index of regulatory sequence elements, and a powerful assay for screening cis-regulatory variants. Like other common diseases, SZ-associated variants are enriched in ENCODE-annotated open chromatin[7, 43, 51, 53]. An effort to identify functional non-coding elements in the brain (PsychENCODE program by NIMH) may complement the ENCODE-annotated chromatin state marks by providing information more relevant to neuropsychiatric disorders, thus facilitating the illumination of more specifi c SZ-risk variants with regulatory potential. However, the accuracy of functional annotation based on physical location in open chromatin is limited by the assay resolution (~600 bp)[40], sequence context-dependent buffering[54], and the lack of disease-relevant cell/tissue types[39]. Empirical testing of the functionality of putative regulatory variants of interest in disease-relevant cell/tissue types thus remains necessary.
Post-transcriptional Regulation
The importance of post-transcriptional regulation, namely mRNA stability and protein translation control, is becoming increasingly appreciated in understanding the dysregulation of synaptic development and function related to neuropsychiatric disorders[55]. Dysregulated synaptic protein synthesis is linked to the abnormal synapse formation, axon arborization, and plasticity in autism[55–57]. In support of this is the widespread and extensive lengthening of 3' UTRs (untranslated regions) that are targeted by miRNAs in the mammalian brain[58]. Targeting the dysregulation of protein synthesis opens up a novel approach for the effective treatment of some neuropsychiatric disorders[55–57, 59].
One of the biological insights from SZ GWAS is the enrichment of noncoding RNAs in top-ranking association hits of PGC (Psychiatric Genomics Consortium) SZ GWAS[6, 7]. A major player is microRNA, small (~22-nt) noncoding RNA that binds to the 3’-UTR of mRNAs, promoting RNA decay and/or repressing mRNA translation (protein synthesis). miRNA dysfunction has been suggested in neurodevelopmental disorders such as autism and SZ[9, 60–65]. Recent SZ GWASs have further strengthened evidence for an etiological role of miRNAs in SZ. Among >100 GWAS-implicated SZ-risk loci[2–7, 66], 24 loci span a total of 33 miRNA genes of which 15 are expressed in the brain (based on BrainSpan-http://www.brainspan.org). Three (MIR124-1, MIR132 and MIR137) are known to regulate neurogenesis, dendritic plasticity, and synaptic function[63, 67–80] and MIR132 also shows reduced expression in SZ postmortem frontal cortex[81, 82]. The predicted (by TargetScan) target genes of these brain-expressed miRNAs are enriched for gene ontology terms associated with neuron development, differentiation, neuron projection, axon guidance, synapse, calcium ion transport, learning and memory, and/or locomotion. There is also a 3-fold enrichment of glutamate receptors among the brain-expressed target genes of these miRNAs. These miRNA-mediated functional gene networks fit well with the known SZ-relevant cellular phenotypes such as reduced synapse density, abnormal circuit connectivity, and synaptic transmission[14–16]. Although most common SZ risk variants or their LD proxies from SZ GWAS may not directly involve the fine regulation of target gene expression, rare genetic variants in miRNA targeting sites may post-transcriptionally tune the expression of genes pathophysiologically important to SZ such as DRD2[32].
Rare regulatory variants conferring a risk for SZ remain to be identified. In this regard, the analytic approach for testing association with rare regulatory variants may need conceptual improvements; for instance, variants in predicted or empirically proven promoters/enhancers may not be simply aggregated together with variants in transcriptional insulators for association tests, because of possible differential effects on the direction of expression. It is clear that gene regulation may play an important role in SZ pathogenesis, but the available eQTL catalogs and ENCODE-based functional annotations have not yet provided power, cellular specificity, or developmental diversity to provide clear mechanistic hypotheses for biological follow-up. It is thus imperative to empirically identify which SZ-risk variants alter chromatin states and ultimately affect gene expression in an experimental model relevant to SZ.
iPSC-Derived Neurons as A Model for Studying Regulatory Variants in Psychiatric Disorders
iPSC-derived neurons are a good model for studying psychiatric disorders for the following reasons: (1) regulatory variants are often cell-type- and developmental stage-specific[37–39], and (2) iPSC neuron differentiation provides an experimental model pathophysiologically relevant to SZ[14, 83–88]. Other alternatives, such as human postmortem brain tissue and genetically-modified model organisms[89–91] have provided insights into SZ pathophysiology, but also have limitations. The postmortem brain is not a living tissue and thus does not capture changes at early neuronal developmental stages[88]. Furthermore, gene expression in postmortem brain is well-known to be confounded by tissue variability and environmental factors[36], and postmortem brain is not amenable to genetic modification. Animal models often do not faithfully reflect the human pathophysiology, especially for brain disorders. Moreover, animal models may not elicit the expected functional impact of human variations[92] because regulatory variants are often species-specific[93]. Although the immune hypothesis remains vital for SZ, which is indeed supported by SZ GWAS findings (the strongest SZ risk locus is at the major histocompatibility complex), after all, schizophrenia is a brain disorder. An abnormal function or expression level of a gene in peripheral blood may predispose an individual to the risk of SZ; however, the phenotypic expression of SZ is closely associated with dysfunction of the brain, manifesting as various abnormal cellular or physiological phenotypes such as reduced functional cortical connectivity[17–20] and reductions in dendritic spine density[17, 18, 21, 22]. iPSC-derived neurons thus provide a unique model to resolve more disease-relevant cellular and molecular phenotype changes as a result of genetic perturbation.
iPSC Generation and Characterization
Most of the commonly-used source tissues or cells for iPSC production are skin biopsies and blood cells, and there are multiple ways of delivering the pluripotent re-programming factors (Oct3/4, Klf4, Sox2, and c-Myc), which have been reviewed extensively elsewhere[94]. The optimal way to generate iPSC lines would be free of virus integration, e.g., by Sendai virus[95], that has been used by the NIMH stem cell center (http://nimhstemcells.org) to establish a resource of iPSC lines for psychiatric research. As iPSC clones can vary substantially, it is necessary to characterize the pluripotency and other biochemical and epigenetic properties of different iPSC clones for the same individual. Besides the confirmation of morphology and positive immunofluorescence staining of pluripotent stem-cell markers (e.g., TRA-1-60, OCT4, NANOG, and SSEA4) (Fig. 2A), full pluripotency of an iPSC line is often confirmed by PluriTest assay (www.pluritest.org)[96] of transcriptome data (Illumina HT-12v4 array) and by the capacity to form embryoid bodies that spontaneously differentiate into the three germ layers (ectoderm, endoderm, and mesoderm) (Fig. 2B). More extensive characterization includes aberrant genetic/epigenetic modification[97], reprogramming-induced point mutations[98] and CNVs[99], or whole-genome sequencing. Because of the technical and financial limitations of scaling-up iPSC production and characterization, generating iPSC lines has often been restricted to small samples, with an emphasis on modeling specific mutations of relatively large effect, e.g., SZ-associated rare CNVs.
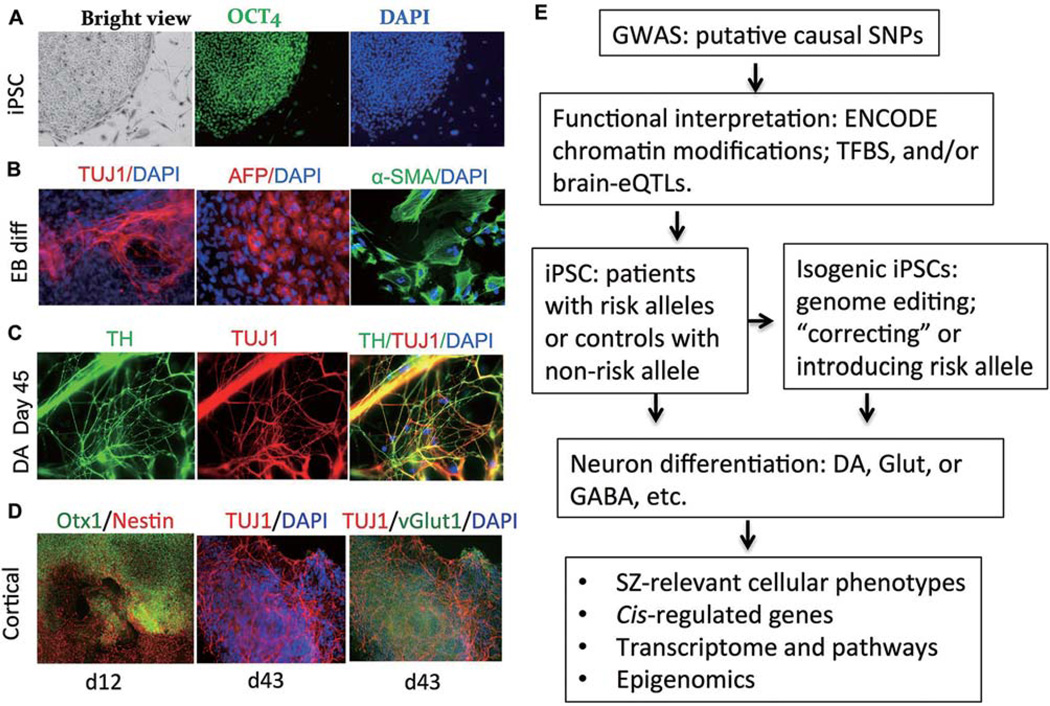
Fig. 2.
Human iPSC-derived neurons as a model for studying regulatory variants of SZ. (A) Human iPSCs characterized by positive immunofluorescence staining for the pluripotent stem cell markers TRA-1-60, OCT4, NANOG, and SSEA4. Only OCT4 staining is shown. DAPI stains the nuclei. (B) Germ layers from embryoid bodies stained positive for TUJ1 (βIII-tubulin), AFP (α-fetoprotein), α-SMA (α-smooth muscle actin), markers specific for ectoderm, endoderm, and mesoderm cells respectively. (C) iPSCs subjected to dopaminergic neuronal differentiation. Neurons are TUJ1+, and DA neurons are TH+. (D) iPSCs subjected to cortical neuronal differentiation. Neuron progenitor cells are nestin+ with cortical identity (Otx1+). Neurons are TUJ1+, and glutamatergic neurons are vGlut1+. (E) Flowchart for the functional characterization of regulatory variants implicated by SZ-GWAS. Photomicrographs for DA neuron differentiation are from Shi et al J Biol Chem, 2014[32].
iPSC-Neuron Differentiation
It is important to determine what would be an appropriate neuronal subtype to derive from iPSCs to model the genetic perturbation of SZ-relevant molecular and cellular phenotypes. iPSCs can be differentiated into multiple major types of neurons (dopaminergic, glutamatergic, and GABAergic)[100] that are relevant to SZ pathophysiology. Dopaminergic neuronal differentiation from iPSCs is the most developed method with relatively high purity[86]. We have achieved ~80% dopaminergic neuronal differentiation efficiency[32] using a floor-plate-based midbrain DA neuronal differentiation method[86]. We observed neurons with DA characteristics, i.e., tyrosine hydroxylase (TH)+ and pituitary homeobox 3 (PITX3)+ at day 30 after plating iPSCs and denser TH+ staining at day 45 (Fig. 2C)[32]. We also found a dynamic expression change of DRD2 that is inversely correlated with the expression of two miRNAs (miRNA-9 and miRNA-326) that target to the 3’-UTRs of DRD2, suggesting a pathophysiologically and developmentally relevant post-transcriptional regulation of DRD2 by both miRNAs[32]. Consistently, we also found an inverse correlation of DRD2 and the two miRNAs in multiple brain regions during brain development[32].
iPSC can also be efficiently differentiated into cortical excitatory neurons, mimicking a process of human cortical development[84, 85]. Given the emerging evidence of dysfunctional frontal cortical development[11–16] and abnormal glutamatergic neurotransmission in SZ[6, 7], iPSC-derived cortical excitatory neurons may be a very important experimental model for studying SZ disease biology. The cortical neurogenesis from iPSCs lasts ~2 months after neuronal induction, which is similar to the ~70-day period of cortical neurogenesis in humans[84]. The cortical neuronal differentiation from iPSCs is reproducible[85]. iPSC-derived neurons include early-born (~day 35) deep-layer Tbr1+/ CTIP2+ neurons and later-born (~day 70) upper-layer Brn2+/Cux1+/Satb2+ neurons, most of which are vGlut1+ (~70%)[84]. We have observed ~100% neuron progenitor cells (NPCs; nestin+) with cortical identity (Otx1/2+) at ~day 12, and a substantial number of vGlut1+ neurons at ~day 43 after neuron induction (Fig. 2D).
The most difficult type of neuron to derive from iPSCs is the GABAergic interneuron. Parvalbumin (PV) GABAergic interneurons have been shown to be relevant to SZ pathogenesis[101]. However, the above-mentioned cortical neuron differentiation procedure does not produce PV+ interneurons[84]. Recently, human pluripotent stem cells (hPSCs) were successfully differentiated into GABAergic interneuron with mature physiological properties along a prolonged intrinsic timeline of up to 7 months, mimicking endogenous human neural development[102]. These neurons express ventral telencephalic GABAergic neuronal lineage markers (ASCL1, DLX1, and DLX5) with increasing expression intensity over time. About 75%–86% of neurons express GABAergic markers (GAD1, SLC32A1, and SLC6A1), and about 53%–78% of neurons express VGAT from 5 to 30 weeks post-differentiation[102].
A common challenge of using iPSC-neurons as a model is the heterogeneity of neuronal culture. There has been a lack of specific cell-surface markers for purifying live neurons[103]. For cortical neurons, although a new method is reported to yield 100% excitatory neurons in a much shorter time than typical cortical neuron differentiation, it requires genetically-modified iPSCs and only ~20% of neurons are vGlut1+[104]. Nonetheless, with 70%–80% purity of dopaminergic neurons, glutamatergic neurons, or GABAergic neurons derived from iPSCs or hPSCs, we believe the iPSC-derived neuronal model will provide an invaluable tool for studying causal molecular mechanisms underlying the genetic findings in SZ. Alternatively, because iPSC can be quickly transformed into NPCs with high purity, and NPCs show transcriptome profiles similar to the developing brain, iPSC-derived NPCs have been proposed as a suitable model for studying the developmental aspects contributing to SZ[88].
CRISPR-Mediated Genome Editing Empowers the Study of Regulatory Variants in iPSC-Neurons
For common disease variants, directly comparing the differential expression between iPSC-neuron cultures of different subjects carrying risk alleles versus non-risk alleles would require a technically and financially prohibitive number of iPSCs. This is because common variants often have small effects and at the same time, there is substantial variation, in particular the variable genetic background, between iPSC lines[103]. Furthermore, although the iPSC-neuron provides a model to directly test regulatory effects relevant to SZ, the associated high cost and labor prevent scaling up. A prominent solution is the emerging genome-editing technology. Genome editing enables the generation of isogenic iPSC-neurons that differ only at the SNP site of interest, a powerful design that can overcome possible confounding effects of variable genetic backgrounds when comparing differences between cells carrying risk versus non-risk alleles[105–113]. With a genome-editing strategy to generate isogenic iPSC-neurons, a workflow combining bioinformatics prediction and empirical iPSC-neuronal disease modelling (Fig. 2E) will help us to link the genotype to cellular phenotypes for causal regulatory variants implicated by SZ GWAS and by future whole-genome sequencing.
Major genome-editing systems include zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN)[105–113] and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) nuclease-mediated genome editing[114–119]. ZFN has the lowest editing efficiency and the most tedious laboratory procedure. TALENs comprise a FokI nuclease domain and DNA-binding domain that can be engineered to recognize specific DNA sequences. TALEN-mediated editing substantially improves the editing efficiency, and presents higher specificity than regular CRISPR/cas9 editing because of the requirement of a dimer of DNA-binding domains to target the specific sequence flanking the variant site to be edited (Fig. 3A). Although an improved version of TALEN editing using GFP (green) and RFP (red) fusion proteins allows rapid screening for edited iPSC clones[108], constructing TALENs is still a tedious procedure (Fig. 3A) and the editing efficiency is often lower than that of CRIPSR/Cas9. CRISPR/Cas9 editing system is noteworthy for its simplicity, high efficiency, and facility for multiplexing[114–119]. With CRISPR/Cas9, a sequence-specific guide RNA (gRNA) leads Cas9 to create a DNA double-strand break at a target site, which in turn triggers DNA damage repair through non-homologous end-joining (NHEJ) or homology-directed repair (HDR). NHEJ results in small insertions or deletions (indels), while HDR introduces precision allele editing. For studying regulatory variants, one could either disrupt the regulatory sequence flanking the risk allele, create a deletion of the putative regulatory sequence using paired gRNAs, or carry out precise risk-allele “correction” with low editing efficiency (<2%) (Fig. 3). Although it has high editing efficiency, the regular CRSIPR/cas9 editing system is reported to often generate off-target editing, which can be monitored by Surveyor nuclease assay[120] or whole-genome sequencing. Improved CRISPR-mediated editing systems, such as modified Cas9 nuclease and Cas9 nickase, can substantially reduce off-target editing thus providing enhanced editing specificity[121], but it often reduces on-target editing efficiency[114]. A better understanding of the biochemical mechanism of each genome-editing system will lead to the improved design of editing tools to increase the editing specificity while retaining the high editing efficiency.
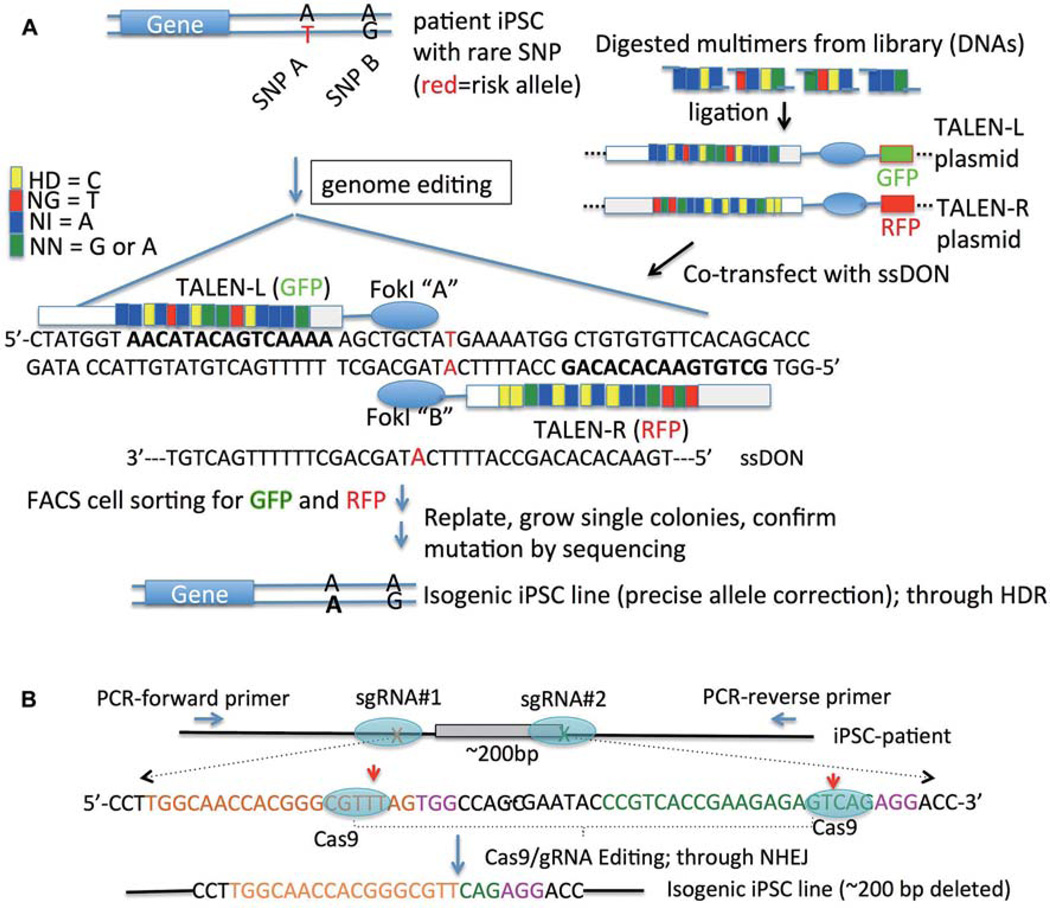
Fig. 3.
Genome editing to create isogenic patient-specific iPSCs. (A) TALEN plasmids are used for precise risk allele correction (T to A for SNP A) through ssDON (single-stranded DNA oligonucleotide)-mediated homology-directed repair. (B) CRISPR/cas9-mediated an exon deletion (~200 bp) using paired sgRNAs (#1 and #2) for targeting. An isogenic iPSC line is generated with ~200 bp DNA being deleted through non-homologous end-joining (NHEJ) of the double-strand breaks of DNA. In each sgRNA, the specific target sequence (20 nt, green or orange) must immediately precede a 5′-NGG adjacent motif (PAM; pink). The red arrow indicates the expected cut site (~3 bp upstream of the NGG PAM sequence) of Cas9 nuclease. Flanking PCR primers can be designed for a rapid screen of iPSC clones carrying the deletion allele.
Genome-modified isogenic iPSC-neurons are thus a powerful approach to compare the functionality of different alleles of single genetic variants on the same genetic background, and using patient-specific iPSCs may further assure a genetic background maximizing the expressivity of a causal variant. However, because of the polygenic nature of complex diseases like SZ, the same single causal variant may still elicit variable functionality in genome-modified isogenic iPSCs derived from different patients with different genetic backgrounds. One way to overcome this limitation would be to characterize the functionality in isogenic iPSC lines derived from more than one patient, and when necessary, combine this with “rescue” experiments to validate the specificity of the observed functional effect.
Conclusion and Outlook
Gene expression regulation contributes substantially to phenotypic variation[28]. Although not sufficient, studying the regulatory effect of a risk allele on cellular phenotypes relevant to SZ is essential for understanding the causal role of a specific regulatory risk variant[122]. Because a simple cellular model such as iPSC-neurons has reduced system “buffering” to genetic or environmental perturbations compared to the whole organism[123], common SZ risk variants with a small population effect size may still elicit moderate or even strong effects on molecular/cellular phenotypes[124–129]. The use of isogenic iPSC-neurons as a disease-relevant experimental model is also expected to enhance the sensitivity of detecting phenotype differences by minimizing the confounding effects of variable genetic backgrounds[123]. Neurons derived from patient-specific iPSCs[14, 83–87] have been used to study SZ-relevant cellular phenotypes such as reduced synaptic density, and abnormal circuit connectivity and synaptic transmission[14–16].
Emerging technology and conceptual innovation will enhance the power of the iPSC-neuron as a model in understanding the disease biology underlying most genetic findings. First of all, defining a sensitive and specific functional assay of regulatory effect on gene expression for neuronal cells is critical. The allele-specific effect on open chromatin as measured by DNaseI HS sites can be an effective functional readout of functional screening for regulatory variants. The most recently developed Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq)[130] provides a much simpler alternative method that requires very few cells for mapping open chromatin, which fits well with studying neuronal cells. Secondly, there is a need for high-throughput genome editing to systematically assay the regulatory effects of a large number of putative regulatory variants. Although high-throughput reporter gene assays can directly examine allelic effects on promoter/enhancer activity of short synthetic regulatory sequences[131, 132], the assayed “function” is not in the context of native genomic architecture. CRISPR/ cas9-mediated genome editing of iPSCs can be used for high-throughput loss-of-function gene screening in regular human cells by sgRNA-guided exon knockout[119] or disruption of non-coding sequences; however, it is still difficult to scale up with a large number of iPSCs, and in particular, the laborious and costly production of iPSC-neurons[123]. The concept represented by the iCRISPR genome editing platform to create compound mutants may have the potential to support high-throughput genetic analysis in iPSCs[133]. Finally, there is still a lack of high-throughput functional assays of neuronal morphology and synaptic properties. Conceptual and technical innovation to develop such functional assays will be fundamental for translating the genetic findings into clinically “actionable” disease biology.
The use of human iPSC-derived cortical neurons enables us to observe molecular and cellular phenotypic alterations more relevant to SZ. However, such an “in-dish” model has limitations in capturing the cortical ultrastructure and synaptic transmission in vivo[123]. Any interesting finding or failure to observe an expected phenotype in iPSC-neurons may thus require cross-platform validation, e.g., in mice[134, 135]. Albeit the limitations mentioned above for mouse models and the different cortical organizations between mice and human brain[135], the basic cellular phenotypes are conserved in the two species and studying abnormal brain development and function in mice has been and remains a powerful approach for modeling genetic perturbations in neuropsychiatric disorders[134]. Indeed, future animal modeling of SZ will benefit from the genetic discoveries from GWAS and from studying iPSC-derived neurons. GWAS provides more specific gene targets for constructing more disease-relevant animal models, while functional genomics in cellular models will inform the causal molecular mechanism of a genetic variant, e.g., whether the risk allele reduces or increases gene expression. This information will thus guide whether knockout (KO) or knock-in (KI) of a target gene in mice is a more appropriate disease model. Furthermore, the current genome-editing technology has been successfully applied to animal disease modeling where one can introduce a specific genetic mutation more efficiently and with higher precision than traditional time-consuming KO/KI animal modeling. Moreover, brain disease modeling is rapidly evolving; for instance, human iPSCs have been recently used to derive 3-D “mini-brains”[136], which may ultimately allow us to understand SZ disease biology in a faithful neurodevelopmental model.
Finally, the identification of specific functional risk variants and their cis-regulated risk genes in broad genomic regions implicated by SZ GWAS and by future whole-genome sequencing studies holds the promise of benefiting SZ pharmacogenomics. New SZ GWAS findings provide an opportunity for developing more effective drug targets as an alternative to the classical antipsychotic drugs that mainly target DRD2[1].
SZ genetic findings may also help to predict which individuals may be more responsive to a particular drug, thus improving the effectiveness of commonly-used antipsychotic drugs. In this regard, iPSC-neurons carrying a specific SZ causal risk variant as a model may serve as a platform for screening drugs that are most effective in a specific subpopulation. Ultimately, the path from SZ genomics to biology will lead to a deeper understanding of the molecular pathogenic mechanisms[122], which will facilitate more precise SZ risk prediction and developing more effective and individualized treatments.
ACKNOWLEDGEMENTS
This review was supported by National Institutes of Health (NIH) Grant R21MH102685 and the North Shore University Health System 2011 Pilot Award.
REFERENCES
- 1.Carlsson A, Lindqvist M. Effect of chlorpromazine or haloperidol on formation of 3methoxytyramine and normetanephrine in mouse brain. Acta Pharmacol Toxicol (Copenh) 1963;20:140–144. doi: 10.1111/j.1600-0773.1963.tb01730.x. [DOI] [PubMed] [Google Scholar]
- 2.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium SPG-WASG. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium SWGotPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennand KJ, Simone A, Tran N, Gage FH. Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry. 2012;17:1239–1253. doi: 10.1038/mp.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennand KJ, Gage FH. Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells. 2011;29:1915–1922. doi: 10.1002/stem.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapoport JL, Giedd JN, Blumenthal J, Hamburger S, Jeffries N, Fernandez T, et al. Progressive cortical change during adolescence in childhood-onset schizophrenia. A longitudinal magnetic resonance imaging study. Arch Gen Psychiatry. 1999;56:649–654. doi: 10.1001/archpsyc.56.7.649. [DOI] [PubMed] [Google Scholar]
- 18.Goghari VM, Rehm K, Carter CS, MacDonald AW., 3rd Regionally specific cortical thinning and gray matter abnormalities in the healthy relatives of schizophrenia patients. Cereb Cortex. 2007;17:415–424. doi: 10.1093/cercor/bhj158. [DOI] [PubMed] [Google Scholar]
- 19.Lewis DA, Gonzalez-Burgos G. Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology. 2008;33:141–165. doi: 10.1038/sj.npp.1301563. [DOI] [PubMed] [Google Scholar]
- 20.Harris KM. Structure, development, and plasticity of dendritic spines. Curr Opin Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- 21.Prasad KM, Goradia D, Eack S, Rajagopalan M, Nutche J, Magge T, et al. Cortical surface characteristics among offspring of schizophrenia subjects. Schizophr Res. 2010;116:143–151. doi: 10.1016/j.schres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harms MP, Wang L, Campanella C, Aldridge K, Moffitt AJ, Kuelper J, et al. Structural abnormalities in gyri of the prefrontal cortex in individuals with schizophrenia and their unaffected siblings. Br J Psychiatry. 2010;196:150–157. doi: 10.1192/bjp.bp.109.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsgodt KH, Sun D, Jimenez AM, Lutkenhoff ES, Willhite R, van Erp TG, et al. Developmental disruptions in neural connectivity in the pathophysiology of schizophrenia. Dev Psychopathol. 2008;20:1297–1327. doi: 10.1017/S095457940800062X. [DOI] [PubMed] [Google Scholar]
- 24.Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Shiino T, et al. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schizophr Bull. 2012;38:552–560. doi: 10.1093/schbul/sbq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 26.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers RA, Casals F, Gauthier J, Hamdan FF, Keebler J, Boyko AR, et al. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet. 2011;7:e1001318. doi: 10.1371/journal.pgen.1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 29.Duan J, Sanders AR, Gejman PV. Genome-wide approaches to schizophrenia. Brain Res Bull. 2010;83:93–102. doi: 10.1016/j.brainresbull.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 31.Duan J, Shi J, Ge X, Dolken L, Moy W, He D, et al. Genome-wide survey of interindividual differences of RNA stability in human lymphoblastoid cell lines. Sci Rep. 2013;3:1318. doi: 10.1038/srep01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi S, Leites C, He D, Schwartz D, Moy W, Shi J, et al. MicroRNA-9 and microRNA-326 regulate human dopamine D2 receptor expression and the microRNA-mediated expression regulation is altered by a genetic variant. J Biol Chem. 2014;289:13434–13444. doi: 10.1074/jbc.M113.535203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, et al. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 37.Nica AC, Ongen H, Irminger JC, Bosco D, Berney T, Antonarakis SE, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul DS, Soranzo N, Beck S. Functional interpretation of non-coding sequence variation: concepts and challenges. Bioessays. 2014;36:191–199. doi: 10.1002/bies.201300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 46.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furey TS, Sethupathy P. Genetics. Genetics driving epigenetics. Science. 2013;342:705–706. doi: 10.1126/science.1246755. [DOI] [PubMed] [Google Scholar]
- 48.Stower H. Gene regulation: from genetic variation to phenotype via chromatin. Nat Rev Genet. 2013;14:824. doi: 10.1038/nrg3622. [DOI] [PubMed] [Google Scholar]
- 49.Kilpinen H, Waszak SM, Gschwind AR, Raghav SK, Witwicki RM, Orioli A, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurano MT, Wang H, Kutyavin T, Stamatoyannopoulos JA. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 2012;8:e1002599. doi: 10.1371/journal.pgen.1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. Autism-related deficits via dysregulated eIF4E–dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3' UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JM, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006767. 206ra138. [DOI] [PubMed] [Google Scholar]
- 60.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 62.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Earls LR, Fricke RG, Yu J, Berry RB, Baldwin LT, Zakharenko SS. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci. 2012;32:14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Earls LR, Bayazitov IT, Fricke RG, Berry RB, Illingworth E, Mittleman G, et al. Dysregulation of presynaptic calcium and synaptic plasticity in a mouse model of 22q11 deletion syndrome. J Neurosci. 2010;30:15843–15855. doi: 10.1523/JNEUROSCI.1425-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weng R, Cohen SM. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development. 2012;139:1427–1434. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- 68.Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, et al. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 2012;8:e1002515. doi: 10.1371/journal.pgen.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, et al. Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fischbach SJ, Carew TJ. MicroRNAs in memory processing. Neuron. 2009;63:714–716. doi: 10.1016/j.neuron.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 72.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ. 2012;19:1573–1581. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. S471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vaccarino FM, Stevens HE, Kocabas A, Palejev D, Szekely A, Grigorenko EL, et al. Induced pluripotent stem cells: a new tool to confront the challenge of neuropsychiatric disorders. Neuropharmacology. 2011;60:1355–1363. doi: 10.1016/j.neuropharm.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carlson GC, Talbot K, Halene TB, Gandal MJ, Kazi HA, Schlosser L, et al. Dysbindin-1 mutant mice implicate reduced fast-phasic inhibition as a final common disease mechanism in schizophrenia. Proc Natl Acad Sci U S A. 2011;108:E962–E970. doi: 10.1073/pnas.1109625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong Z, Peng J, Guo S. Stable Gene Silencing in Zebrafish with Spatiotemporally Targetable RNA Interference. Genetics. 2013 doi: 10.1534/genetics.112.147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jeong JY, Einhorn Z, Mercurio S, Lee S, Lau B, Mione M, et al. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc Natl Acad Sci U S A. 2006;103:5143–5148. doi: 10.1073/pnas.0600337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brennand KJ, Landek-Salgado MA, Sawa A. Modeling Heterogeneous Patients With a Clinical Diagnosis of Schizophrenia With Induced Pluripotent Stem Cells. Biol Psychiatry. 2014;75:936–944. doi: 10.1016/j.biopsych.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seki T, Yuasa S, Fukuda K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc. 2012;7:718–728. doi: 10.1038/nprot.2012.015. [DOI] [PubMed] [Google Scholar]
- 96.Muller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 100.Wright R, Rethelyi JM, Gage FH. Enhancing induced pluripotent stem cell models of schizophrenia. JAMA Psychiatry. 2014;71:334–335. doi: 10.1001/jamapsychiatry.2013.4239. [DOI] [PubMed] [Google Scholar]
- 101.Nakazawa K, Zsiros V, Jiang Z, Nakao K, Kolata S, Zhang S, et al. GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology. 2012;62:1574–1583. doi: 10.1016/j.neuropharm.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soldner F, Jaenisch R. Medicine. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2012;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2012;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013 doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 124.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal lari R, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller CL, Haas U, Diaz R, Leeper NJ, Kundu RK, Patlolla B, et al. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014;10:e1004263. doi: 10.1371/journal.pgen.1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Spieler D, Kaffe M, Knauf F, Bessa J, Tena JJ, Giesert F, et al. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kulzer JR, Stitzel ML, Morken MA, Huyghe JR, Fuchsberger C, Kuusisto J, et al. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am J Hum Genet. 2014;94:186–197. doi: 10.1016/j.ajhg.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, et al. Massively parallel functional dissection of mammalian enhancers in vivo . Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 135.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]