Figure 3.
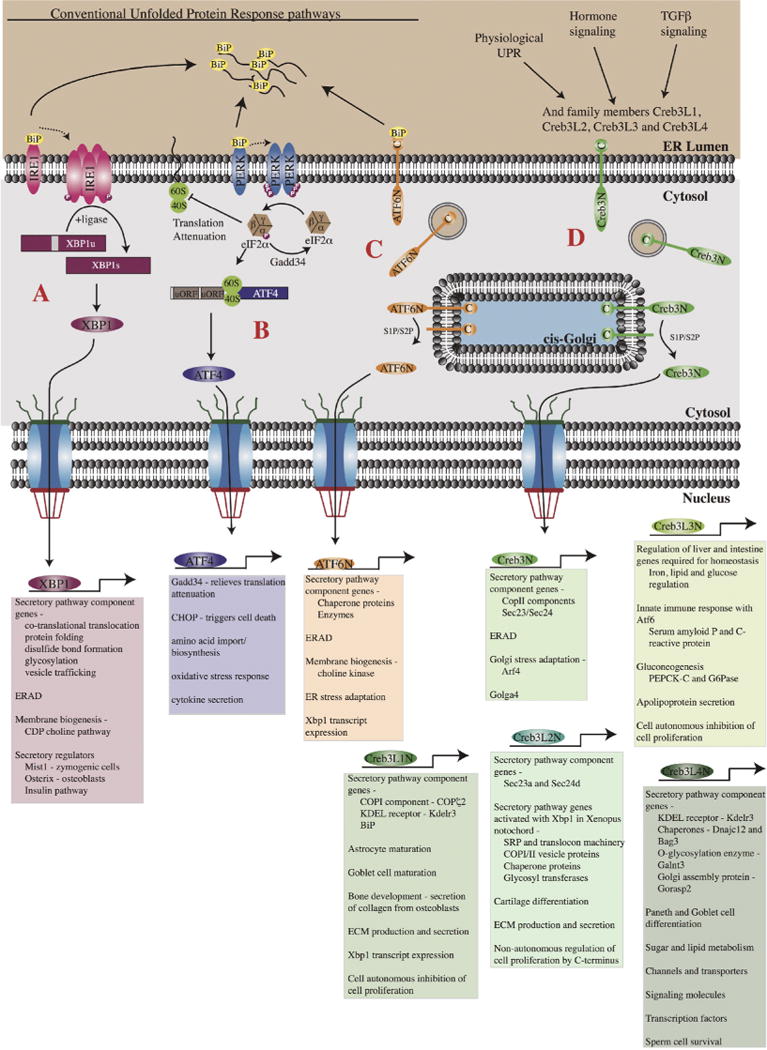
Regulation and function of the bZip transcription factors that upregulate secretory capacity during the UPR and physiological ER stress. (A). The Ire-1 kinase is held inactive by the binding of the chaperone protein BiP. Accumulation of unfolded proteins leads to the release of BiP allowing it to oligomerize and autophosphorylate thereby activating the RNAse domain. The primary substrate for Ire-1 in mammalian cells is the Xbp1 mRNA, which gets spliced by Ire-1. Following translation, the active Xbp1 transcription factor translocates to the nucleus where it activates target genes involved in the secretory pathway, membrane biogenesis, ERAD, and secretory cell differentiation. (B). PERK also binds to BiP and is only activated following the release of BiP in the presence of unfolded proteins. BiP release leads to dimerization of PERK and the subsequent phosphorylation of eIF2α, leading to translational attenuation. This event also leads to the preferential translation of the ATF4 mRNA. ATF4 then functions in the nucleus to regulate genes involved in secretory activity as well as UPR dependent cell death. (C). ATF6 is an ER membrane bound transcription factor that during the UPR traffics to the Golgi and is cleaved by the Site-1 and Site-2 proteases releasing its N-terminal bZip domain. The active ATF6 transcription factor then translocates to the nucleus where it regulates genes involved in protein folding, membrane biogenesis and ERAD. (D). The Creb3 transcription factors are also ER-membrane bound and similar to ATF6 undergo cleavage by the Site-1 and Site-2 proteases. Unlike ATF6 (or the other canonical members of the UPR), Creb3 proteins do not bind BiP but are instead activated by physiological changes in secretory demand. Following cleavage, the N-terminal transcription factor domain translocates to the nucleus to activate gene transcription. Creb3 family members are largely involved in regulating the protein machinery of the early secretory pathway.
