Summary
Inhibition of host-encoded targets, such as the cyclophilins, provides an opportunity to generate potent, high barrier to resistance antivirals for the treatment of a broad range of viral diseases. However, many host-targeted agents are natural products which can be difficult to optimize using synthetic chemistry alone. We describe the orthogonal combination of bioengineering and semisynthetic chemistry to optimize the drug-like properties of sanglifehrin A, a known cyclophilin inhibitor of mixed non-ribosomal peptide/polyketide origin in order to generate the drug candidate NVP018 (formerly BC556). NVP018 is a potent inhibitor of HBV, HCV and HIV-1 replication, shows minimal inhibition of major drug transporters and has a high barrier to generation of both HCV and HIV-1 resistance.
Introduction
Treatment of chronic viral diseases by direct inhibition of viral targets frequently leads to rapid development of virally-encoded resistance. Therapies targeted to host proteins involved in the viral life-cycle offer an opportunity to both raise the barrier to generation of resistance and generate antivirals able to treat a broad range of viral diseases. However, this has been hindered in the past by the relative rarity of targets essential to the virus, but non-essential to the host, and the inherent complexity of discovering and developing drugs that effectively target these proteins.
From the 1940s to 2007, 73% of the 155 small molecules approved as anticancer drugs were of natural origin, either directly or derived1. As opposed to this, few natural products had been approved for use in antiviral therapy, although they were the inspiration for antiviral nucleoside analogues2.
Many bacterial natural products (NPs), including the well-described polyketide class, have been shown to bind to and inhibit mammalian cellular proteins, including viral host-encoded targets such as cyclophilins, v-ATPases3, CRM14 and Hsp905. They can be produced at low cost of goods by fermentation and frequently the lead compound has both excellent potency against the target and good cellular penetration. However, natural products often display polypharmacology and have less than optimal mammalian pharmacokinetics and physicochemical properties.
The tools available for optimizing NPs are ever increasing. Specifically, bioengineering has been proposed as a way to reinvigorate natural products drug discovery6,7. In the past, semisynthetic approaches have been the most frequently used route to improve the drug-like properties of a natural product hit. However, the available semi synthetic options are pre-determined by the array of functional groups on the NP. In contrast, bioengineering options are pre-determined by the biosynthetic pathway. The potential changes are thus orthogonal to those available to semisynthesis.
Whole genome sequencing is now straightforward and easily affordable, and for NP classes such as the modular polyketide synthases (PKSs), knowledge of the DNA sequence encoding the biosynthetic gene cluster enables rapid understanding of gene product function. When combined with improved techniques for DNA transfer and the rapid targeted alteration of biosynthetic genes this provide a powerful platform for focused drug discovery efforts with the aim of improving drug-like properties, pharmacokinetics and reducing off-target effects. These bioengineering techniques are readily combined with semisynthesis to identify molecules with further improved properties. In particular, inactivation of precursor pathways can allow mutasynthesis, the process of feeding a synthetic analogue of the precursor which is then incorporated, biosynthetically, into the final molecule8,9. This enables a combinatorial element to bioengineering.
Cyclophilins are a class of peptidyl-prolyl isomerases, proteins which catalyse the cistrans isomerization of the peptide bond preceding prolyl residues. Knockout studies in several species, including mice and human cells, confirm that they have limited or no effect on cellular growth and survival10–13. However, cyclophilins recruited from host cells have been shown to have essential roles in many viral life-cycles. Initially, cyclophilin A was shown to be incorporated into HIV-1 virions14,15, involved in viral replication, and its expression level in patients related to the speed of progression to AIDS16. Cyclophilin A isomerase activity, and potentially other cyclophilins such as B and cyclophilin 40, have been shown to be required for HCV replication17–19. Other viruses where cyclophilin involvement has been implicated in their life-cycle, or where cyclophilin inhibitors have shown inhibitory activity, include Vaccinia virus20, West Nile virus, Dengue virus, Yellow fever virus21, Hepatitis B virus22, Human papilloma virus23, Cytomegalovirus24, SARS coronavirus25, Japanese encephalitis virus26 and Influenza A27.
We now describe the use of combined bioengineering and semisynthetic approaches to optimize the drug-like properties of sanglifehrin A, a NP cyclophilin inhibitor, to generate NVP018 (formerly BC556). Preclinical analysis reveals NVP018 to be a molecule displaying in vitro inhibition of HBV and HCV and potent in vitro and oral in vivo inhibition of HIV-1.
Results
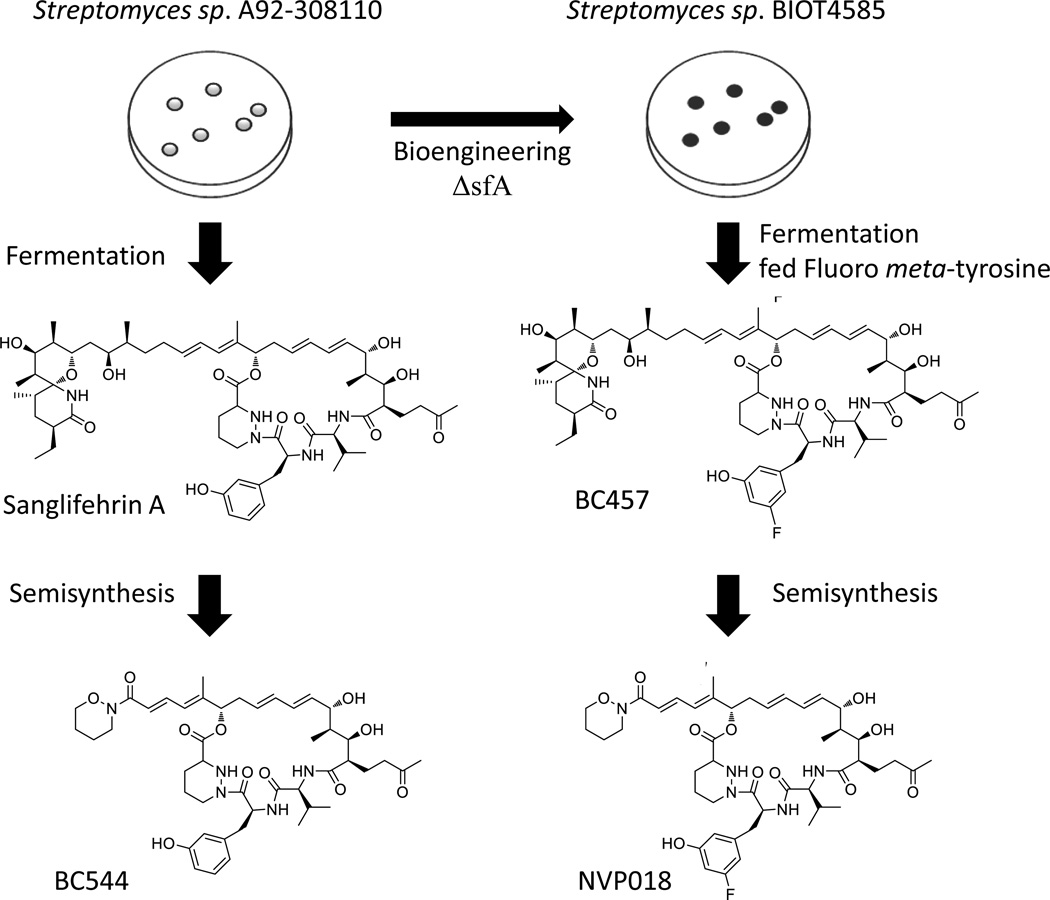
Bioengineering and mutasynthesis of Streptomyces sp. A92-308110
Previous semisynthetic derivatization to replace the spirolactam fragment of sanglifehrin A led to generation of the sangamides, molecules with improved solubility, potency and selectivity28. This approach moved the immunosuppressive NPs from tool compounds appropriate for chemical genetics to being compounds that could reasonably be pursued as therapeutic agents. We then used bioengineering to make further alterations to the macrocyclic portion of the molecule, changes which could not have otherwise been done without complex total synthesis. The major aim of this work was to further improve the drug-like properties of the final molecule by specific manipulation of the parts of the molecule involved in cyclophilin binding.
The gene cluster required for biosynthesis of sanglifehrin A has been published30. Generation of the sanglifehrins requires thirteen typical PKS extension modules, and three NRPS (non-ribosomal peptide synthetase) modules incorporating l-valine, meta-l-tyrosine and l-piperazic acid. In addition, a crotonyl-CoA reductase/carboxylase (CCR) is present for generation of the PKS starter unit (2E)-2-ethylmalonyl-CoA, an iterative type I PKS is required for the supply of the unique (2S)-2-(2-oxo-butyl)malonyl-CoA extender unit, and a phenylalanine hydroxylase produces the NRPS extender unit meta-l-tyrosine from l-phenylalanine.
Analysis of the published crystal structures of sanglifehrin A31 and an analogue with the spirolactam fragment removed32, suggested that manipulation of the electronics on the meta-l-tyrosine moiety might lead to improved binding to cyclophilins. To achieve this, we inactivated sfaA, the gene encoding the phenylalanine hydroxylase responsible for meta-l-tyrosine biosynthesis. Deletion of sfaA was carried out by homologous recombination, following conjugation of a plasmid containing the appropriate deletion construct into S. sp. A92-308110 using standard techniques33 and led to the strain BIOT-4585. Due to the lack of meta-l-tyrosine production BIOT-4585 did not produce sanglifehrin A unless fed with meta-l-tyrosine or an analogue.
Synthesis and selection of NVP018
A screen was then carried out by feeding a selection of appropriate meta-l-tyrosine analogues to BIOT-4585 in order to determine its ability to generate sanglifehrin analogues (see figure 1). Fermentations with those analogues successfully accepted as substrates were scaled up to allow isolation of new sanglifehrins. The antiviral activity and ADME properties of each analogue were analysed, and the best of the analogues were subjected to semisynthesis to yield the corresponding sangamides as described previously28. The resulting sangamides were again analysed for antiviral activity and ADME properties, and from this NVP018 was chosen as a potential drug candidate (data not shown). NVP018 is derived from a sanglifehrin resulting from incorporation of 5-fluoro-meta-l-tyrosine. Semisynthesis replaced the spirolactam containing portion of the sanglifehrin with a tertiary hydroxamic amide moiety (table S1). Of the sangamides tested, NVP018 had the best combination of properties (table 1), including more potent inhibition of CypA PPIase activity and inhibition of HCV replicons.
Figure 1.
Overview of (bio)synthetic medicinal chemistry route to generate BC544 and NVP018
Table 1.
Comparison of cyclophilin inhibition, HCV NS5A-cyclophilin A complex disruption and genotype 1b replicon inhibition for cyclosporin and sanglifehrin analogues
| CypA PPIase IC50 (nM) |
NS5A-CypA disruption IC50 (µM) |
HCV Huh5.2 Gt1b replicon EC50 (nM) |
HCV Gt1b replicon CC50 (µM) |
|
|---|---|---|---|---|
| CsA | 9.7 ± 1.30 | 0.98 ± 0.08 | 306 ± 141 | 4.4 |
| Alisporivir | 0.8 ± 0.05 | 0.24 ± 0.02 | 96 ± 23 | 11.2 |
| BC544 | 2.7 ± 0.2 | 0.36 ± 0.03 | 125 ± 22 | >100 |
| NVP018 | 0.3 ± 0.03 | 0.10 ± 0.01 | 33 ± 10 | >100 |
NVP018 has reduced off target inhibition of drug transporters
One of the issues with cyclosporin-based cyclophilin inhibitors is off-target inhibition of transporter proteins. Alisporivir, for example, inhibits multiple transporter proteins, including OATP1B1, OATP1B3 and MRP2, which are involved in bilirubin transport. It is this inhibition which is now thought to be the mechanism for the dose limiting hyperbilirubinaemia34. These compounds also have drug-drug interaction concerns through inhibition of xenobiotic transporters such as Pgp and BSEP35,36. Comparison of the inhibition of these transporters using vesicular transport or uptake transporter inhibition assays, and in vitro inhibition of the xenobiotic transporters, Pgp and BSEP revealed that whilst alisporivir and CsA potently inhibited many of them, BC544 (figure 1, previously stated as compound 3o in Moss et al., 2011) and NVP018 showed minimal or no inhibition (see table 2). To test the effect of NVP018 on bilirubin levels, we then dosed three mice with up to 250 mg/kg/day NVP018 for 7 days. In all cases, almost no increase was seen in either bilirubin or liver enzymes when measured at the end of the study (see supplementary information, table S2).
Table 2.
Comparison of solubility, plasma protein binding and transporter inhibition
| Solubility in PBS pH7.4 (uM) |
Human PPB (fraction unbound) |
OATP1B1 IC50 (µM) |
OATP1B3 IC50 (µM) |
MRP2 IC50 (µM) |
Pgp IC50 (µM) |
BSEP IC50 (µM) |
|
|---|---|---|---|---|---|---|---|
| CsA | 36 | <1% | 0.85 | 0.13 | 4.1 | 0.73 | 0.46 |
| Alisporivir | 15 | <4% | 0.45 | 0.19 | ~16 (>49%) | 0.72 | 0.18 |
| BC544 | 330 | 28% | 10.97 | 5.40 | >50 | >50 | 29.3 |
| NVP018 | 140 | 22% | 4.31 | 1.75 | >50 | >50 | 12.3 |
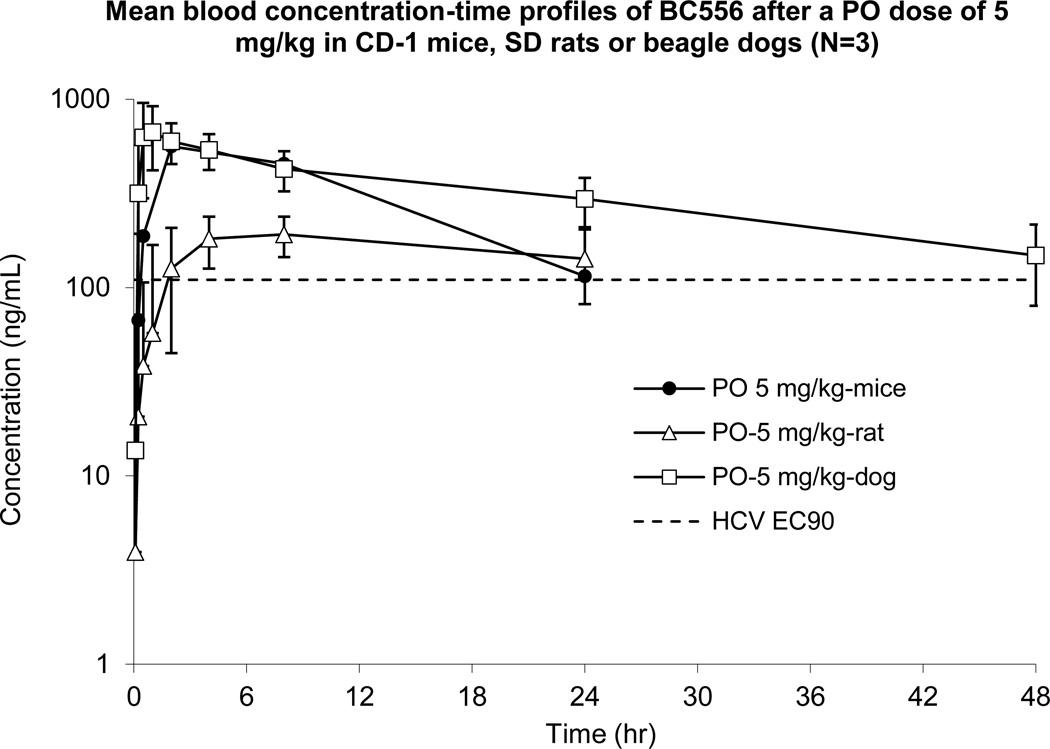
NVP018 displays in vivo pharmacokinetics suitable for once daily dosing
To confirm bioavailability following oral dosing, a single oral dose of 5 mg/kg was given to groups of mice, rats and dogs. In all cases, blood concentrations were maintained above the HCV EC90 for 24–48 h post dose (figure 2). Liver levels were found to be 3–6 fold higher than blood levels, and up to 230 fold higher than plasma levels, revealing further concentration within the target organ for treatment of hepatic disease (see supplementary information, table S3). Analysis of other organs in rats, including brain, heart and lungs revealed much lower exposure following oral dosing as compared to the blood and liver, probably due to first pass extraction by the liver.
Figure 2.
Mean blood concentration-time profiles of NVP018 following a single po dose of 5 mg.kg−1 to CD-1 mice, SD rats or beagle dogs
NVP018 is an inhibitor of HCV replication in vitro
Until recently the standard of care treatment for chronic HCV infection has been a combination of interferon and ribavirin for 6–12 months. Use of recently approved high barrier to resistance drugs, such as the nucleotide polymerase inhibitor (NPI) sofosbuvir leads to high levels of viral clearance37,38. Cyclophilin inhibition is one of the few mechanisms of action of HCV inhibition which consistently shows a high barrier to resistance, and in Phase 2 trials, some patients were clear of the virus after being treated for 24 weeks with 1000 mg/day alisporivir monotherapy39. Alisporivir, a non-immunosuppressive semisynthetic derivative of cyclosporin A, is a potent cyclophilin inhibitor in Phase III trials for the treatment of HCV infection. NVP018 could provide a suitable alternative backbone for use in an interferon-free combination. Comparison of the inhibitory potential of NVP018 with the semisynthetic sangamide, BC544 and alisporivir against HCV replicons revealed that NVP018 was much more potent in all assays, and was similarly active against all genotypes tested (see table 3). Analysis of the effect of increasing levels of either human serum or FCS also confirmed that there was no effect on antiviral potency with addition of up to 40% serum (see supplementary information table S4).
Table 3.
EC50 of cross genotype HCV replicon inhibition and IC50 of HIV-1 inhibition. IC50 of HepG2 cells stably replicating HBV following treatment with DMSO, tenofovir, BC544, NVP018 and Alisporivir and test/control value after 3 days of Huh7 cells following transfection with HBV DNA in the presence of DMSO, tenofovir, BC544, NVP018 and Alisporivir at 2 µM
| HCV | HIV-1 | HBV | ||||||
|---|---|---|---|---|---|---|---|---|
| HCV Huh7 Gt1a replicon EC50 (nM) |
HCV Huh7 Gt1b replicon EC50 (nM) |
HCV Huh7 Gt2a replicon EC50 (nM) |
HIV-1 HeLa cells IC50 (nM) |
HBV Huh7 stable HBV DNA IC50 (nM) |
HBV Huh7 stable HBsAg IC50 (nM) |
HBV Huh7 stable HBeAg IC50 (nM) |
HBV HepG2 transfected HBsAg T/C d3 at 2uM (%) |
|
| CsA | 198 ± 13 | 137 ± 4 | 455 ± 37 | 5300 | N/A | N/A | N/A | N/A |
| Alisporivir | 98 ± 11 | 76 ± 5 | 102 ± 8 | 1500 ± 240 | >10000 | >10000 | >10000 | 18 |
| BC544 | 63 ± 4 | 58 ± 5 | 89 ± 6 | 310 ± 20 | 610 | 600 | 280 | 9.6 |
| NVP018 | 18 ± 2 | 13 ± 1 | 21 ± 3 | 130 ± 15 | 950 | 520 | 350 | 8.8 |
To compare the barrier to resistance of NVP018 in HCV, Huh7-Con1 cells were serially passaged for 7 weeks in the presence of 300 µg/mL G418 and increasing concentrations of NVP018 or alisporivir. Colonies were amplified, cellular RNA extracted and analyzed by RT-qPCR. For all alisporivir-resistant replicons, only the previously described and clinically relevant D320E mutation was consistently found in all clones analyzed. In the case of NVP018 resistant replicons, all clones analyzed contained the same five mutations: L27I, R318W, D320E, S370P and C446S. When NVP018 was tested against separate replicons containing individual mutations from this list, only the D320E mutation was shown to confer more than 2-fold resistance alone. The combination of all five mutations only led to ~4-fold resistance compared to the wild-type.
NVP018 is an inhibitor of HBV replication in vitro
There have been previous reports that cyclophilin inhibitors such as cyclosporin A have an inhibitory effect on HBV replication40. We therefore tested NVP018 in two in vitro models to look for inhibitory activity. The first involved transfecting Huh7 cells with HBV DNA (8 µg) in the presence of 2 µM NVP018, alisporivir or DMSO. After days 0, 1, 2 and 3, amounts of de novo HBV proteins HBsAg and HBeAg present in cell extracts or supernatants were quantified by ELISA, in triplicate. Data is shown in Table 3 and revealed that NVP018 and alisporivir potently inhibited HBV replication at this concentration. An additional assay was run using HepG2 cells stably expressing replicating HBV. These were treated with DMSO, NVP018 or alisporivir at three concentrations (0.1, 1 and 10 µM). The effect on HBV replication was monitored by treating HepG2 cells with test article at the relevant concentration for 3 days, removing the supernatant, retreating and analyzing after an additional 48 and 72 h by HBsAg ELISA, HBeAg ELISA and for HBV DNA, in triplicate, in both cell extracts and supernatants. This assay confirmed a dose dependent effect of NVP018 against HBV replication at the final time point.
NVP018 is an inhibitor of HIV-1 replication in vitro
Although no cyclophilin inhibitors are currently in clinical development for treatment of HIV-1 infection, alisporivir was originally developed as an inhibitor of HIV-1 replication, and showed a mean ~1 log drop in viral load following 14 days dosing in HIV/HCV co-infected patients41. We therefore tested NVP018 in comparison to BC544 and alisporivir in a CD4+ HeLa HIV-1 inhibitory FACS assay to see if improvements in inhibition of in vitro HCV replicons carried through to HIV-1 inhibitory assays. NVP018 was found to be both more potent as regards HIV-1 EC50 (see table 3) and in terms of maximal viral inhibition, which was seen to reach >95% inhibition with sanglifehrin analogues including BC544 and NVP018 but only 60% with cyclosporin A and alisporivir. This data was similar when repeated in Jurkat cells, activated CD4+ peripheral blood T-lymphocytes and macrophages. One of the concerns with development of alisporivir for treatment of HIV-1 was the presence of natural isolates with the H86Q alisporivir resistant genotype22. NVP018 was tested against both lab generated and natural H86Q isolates, and was found to be active in both cases.
NVP018 is a potent inhibitor of HIV-1 replication in vivo
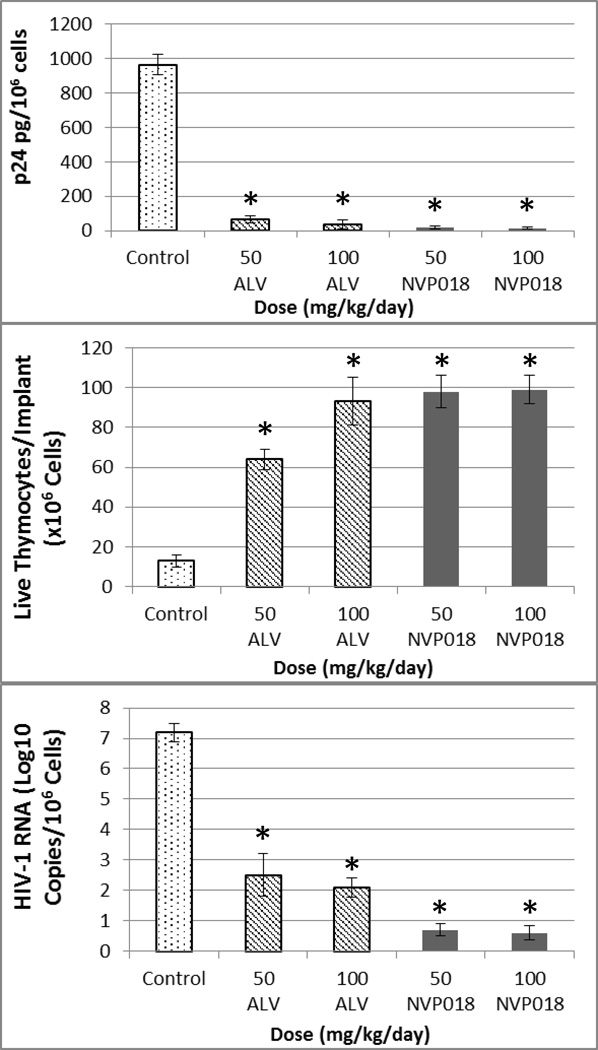
As NVP018 had shown both potent in vitro HIV-1 inhibition and oral bioavailability, its effect on HIV-1 viral replication in the SCID-hu Thy/Liv mouse model was assessed. This model has been well validated against other standard HIV therapies and is thought to be a highly reproducible model with relevance to clinical effect29.
To enable comparison with data from previous studies with approved agents in the model, NVP018 or alisporivir were dosed orally bi-daily for 14 days, starting one day before inoculation with the NL4.3 virus, at 50 mg/kg/day or 100 mg/kg/day. Administration of NVP018 gave >6 log10 reductions in HIV-1 RNA, and was more potent at all concentrations tested than alisporivir at protecting thymocytes, and in reducing HIV-1 RNA and p24 levels (see figure 3). No overt signs of toxicity were seen in any of the mice at any dose level. The potency of NVP018 compares well with data previously published by Stoddart et al. as the >6 log10 HIV-1 RNA log drop seen following NVP018 dosing was more pronounced than for approved therapies such as efavirenz, atazanavir, emtricitabine and nevirapine, which showed 1–4 log10 viral RNA reductions after 21 days bi-daily dosing at 100 mg/kg.
Figure 3.
Comparison of implant P24 levels, T-lymphocytes and HIV-1 RNA in the SCID-hu Thy/Liv mouse model, following bi-daily doses of either 50 or 100 mg.kg−1 alisporivir (ALV) or NVP018 for two weeks. *=p≤0.05, compared with untreated mice
Discussion
The cyclophilin inhibitor NVP018 was generated using a combination of semisynthetic and bioengineering methods. These are frequently used independently, but in the past have been less commonly used together42,43. This enabled a combinatorial approach to natural product drug discovery and led to the selection of NVP018, a compound with more potent cyclophilin inhibition, antiviral potency versus HBV, HCV and HIV-1, and reduced off-target transporter inhibition as compared to the parent compound, sanglifehrin A. A fluorine was introduced into the compound by mutasynthesis, a method which has been successful in introducing this substitution pattern in a less invasive way than semisynthetic chemistry44,45,46,47,48.
Second generation cyclophilin inhibitors, such as NVP018, have the potential for therapeutic application in a number of areas, in particular chronic viral infection. This is due to their potency, high barrier to resistance, activity on multiple steps of the viral life-cycle and oral bioavailability. It is also worth noting the recent publications which suggest that these compounds might also have additional impact on Interferon Regulatory Factors, which have long been implicated in the escape of certain viruses, such as HCV and HIV, from the immune system49,50. Whilst cyclophilin inhibitors do not exhibit inhibition of all viruses, they have been shown to have activity against a wide variety of both RNA viruses (eg Coronaviruses, HIV and HCV) and DNA viruses (HCMV, HBV, VZV). It is therefore tempting to speculate that cyclophilin inhibitors may be a step in the direction of agents with broad antiviral activity51.
We also anticipate that in the era of rapid genome sequencing and gene synthesis, bioengineering applications will continue to become more amenable and offer a more flexible route to improve natural products with potent inhibition of cellular targets, but with less than optimum drug-like properties. We hope that in the future, bioengineering and semisynthesis will be used more frequently in combination to optimize natural products with an aim to select candidates for clinical development.
Experimental Procedures
Compounds
BC544 was synthesized according to the method published in Moss et al., 2012 (compound 3o therein). Alisporivir (DEBIO-025/DEB025) was synthesized according to the method published in US6,927,208 from cyclosporin A (>99%) purchased from LC Labs, Woburn, MA, USA.
Genetic engineering of Streptomyces sp. A92-308110
In frame deletion of the sfaA gene, coding for the tyrosine hydroxylase, was carried out using double recombination. The recombinant strain generated, BIOT-4585, could then be fed (S)-methyl 2-amino-3-(3-fluoro-5-hydroxyphenyl) propanoate hydrochloride salt and grown under appropriate fermentation conditions to generate BC457. Further details are available in the supplementary information.
Semisynthesis of NVP018 from BC457
NVP018 was synthesized in three steps from BC457 using the same protocol as for compound BC544. Briefly BC457 was treated via modified Sharpless dihydroxylation conditions to form a diol which was subsequently cleaved to generate an allylic aldehyde by the action of sodium periodate. Finally the resultant allylic aldehyde was coupled with a suitable amide under Horner-Wadsworth-Emmons coupling conditions.
ADME Compound analysis
Solubility analysis, plasma protein binding, intravenous and oral pharmacokinetics and toxicology were carried out using standard methods at Shanghai Chempartner, Shanghai and efflux assays were carried out at Solvo Biotechnology, Hungary or Cyprotex, UK. Further details are available in the supplementary information.
ELISA Analysis of CypA-NS5A Interactions
Test articles were tested for their capacities to block the interaction between CypA and HCV NS5A by ELISA. Specifically, we produced and purified recombinant GST, GST-CypA and Con1 NS5A-His proteins and conducted ELISA. Nunc MaxiSorb 8-well strip plates were coated with GST or GST-CypA for 16 h at 4 °C and blocked. Recombinant NS5A-His (1 ng/mL) was added to wells in 50 µL of binding buffer (20 mM Tris pH 7.9, 0.5 M NaCl, 10 % glycerol, 10 mM DTT and 1 % NP-40) for 16 h at 4 °C. Captured NS5A-His was subsequently detected using mouse anti-His antibodies (1 µg/mL) (anti-6×His, Clontech) and rabbit anti-mouse-horseradish peroxidase phosphatase (HRP) antibodies (1:1000 dilution). To test the effect of test articles on the CypA-NS5A complex formation, increasing concentrations of each test article were added to GST-CypA together with recombinant NS5A. All experiments were conducted twice. Data (triplicate) are presented (IC50).
PPIase Inhibition Analysis
The inhibition of the PPIase activity of CypA was used to compare the inhibitory potential of sanglifehrin derivatives as an indication of their binding affinity to CypA. The PPIase activity of recombinant CypA, produced by thrombin cleavage of GST-CypA, is determined by following the rate of hydrolysis of N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide by chymotrypsin. Chymotrypsin only hydrolyzes the trans form of the peptide, and hydrolysis of the cis form, the concentration of which is maximized by using a stock dissolved in trifluoroethanol containing 470 mM LiCl, is limited by the rate of cis-trans isomerization. CypA was equilibrated for 1 hour at 5 °C with selected sanglifehrin derivatives using a drug concentration range from 0.1 to 20 nM. The reaction was started by addition of the peptide, and the change in absorbance was monitored spectrophotometrically at 10 data points per sec. The blank rates of hydrolysis (in the absence of CypA) were subtracted from the rates in the presence of CypA. The initial rates of the enzymatic reaction were analyzed by first-order regression analysis of the time course of the change in absorbance. All sanglifehrin derivatives exhibited anti-PPIase activity that correlated well with their capacities to prevent CypA-NS5A interactions analyzed by ELISA or pulldown.
HCV Antiviral Assay Huh 5-2
Anti-HCV assay in Huh 5-2 cells was performed by seeding 6.5×103 cells per well in a tissue culture treated white 96-well view plate (Packard, Canberra, Canada) in Dulbecco’s modified essential media (DMEM) supplemented with 250 µg/ml G418. Following incubation for 24 h at 37 °C (5 % CO2) medium was removed and 3-fold serial dilutions in complete DMEM (without G418) of the test compounds were added in a total volume of 100 µl. After 4 days of incubation at 37°C, cell culture medium was removed and luciferase activity was determined using the luciferase assay system (Promega, Leiden, The Netherlands); the luciferase signal was measured using a Safire2 (Tecan, Switzerland). Relative luminescence units were converted to percentage of untreated controls. The 50 % effective concentration (EC50) was defined as the concentration of compound that reduced the luciferase signal by 50%.
Cytostatic Assay
For the assessment of the potential cytostatic effect of the evaluated inhibitor, cells were seeded at a density of 6.5×103 cells per well of a 96-well plate in complete DMEM. Serial dilutions of the test compounds in complete DMEM were added 24 h after seeding. Cells were allowed to proliferate for 3 days at 37 °C, after which the cell number was determined by means of the (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)/phenazinemethosulfate (MTS/PMS) method (Promega). The CC50 (value derived from the dose-response curve) represents the concentration at which the metabolic activity of the cells would be reduced to 50 % of the metabolic activity of untreated cells. The selectivity index (SI), indicative of the therapeutic window of the compound, was calculated as the CC50/EC50.
HCV Replicon antiviral assays
The replicon cells (subgenomic replicons of genotype 1a (H77), 1b (Huh7) and 2a (JFH-1)) were grown in DMEM, 10 % fetal bovine serum (FBS), 1 % penicillin-streptomycin (pen-strep), 1 % glutamine, 1 % non-essential amino acids, 250 µg/ml G418 in a 5% CO2 incubator at 37 °C. All cell culture reagents were purchased from Mediatech (Herndon, VA). The replicon cells were trypsinized and seeded at 5 × 103 cells per well in 96-well plates with the above media without G418. On the following day, the culture medium was replaced with DMEM containing compounds serially diluted in the presence of 5 % FBS. The cells containing the HCV replicon were seeded into 96-well plates and test articles were serially diluted with DMEM plus 5 % FBS. The diluted compound was then applied to appropriate wells in the plate. After 72 hours incubation at 37 °C, the cells were processed. The intracellular RNA from each well was extracted with an RNeasy 96 kit (Qiagen). The level of HCV RNA was determined by a reverse transcriptase-real time PCR assay using TaqMan® One-Step RT-PCR Master Mix Reagents (Applied Biosystems, Foster City, CA) and an ABI Prism 7900 sequence detection system (Applied Biosystems). The cytotoxic effects were measured with TaqMan® Ribosomal RNA Control Reagents (Applied Biosystems) as an indication of cell numbers. The amount of the HCV RNA and ribosomal RNA were then used to derive applicable EC50 values (concentration that would inhibit the replicon replication by 50%).
In vitro assays for assessment of HBV antiviral activity
HepG2 cells stably replicating robustly HBV were treated with DMSO, tenofovir, BC544, NVP018 and ALV at 3 concentrations (0.1, 1 and 10 µM). Effect on HBV replication was monitored by treating HepG2 cells with test article at the relevant concentration for 3 days, removing the supernatant, retreating and analyzing after an additional 72 hours by HBsAg ELISA, HBeAg ELISA and HBV DNA in triplicates in cell extracts. These data were used to generate test/control values which were plotted against the test article concentration and used to calculate IC50 values.
Huh7 cells were transfected with HBV DNA (8 µg) in the presence of DMSO, tenofovir, BC544, NVP018 and ALV (alisporivir) at 2 µM. After day 3, amounts of de novo HBsAg protein present in cell extracts or supernatants was quantified by ELISA in triplicate. This was used to calculate a test/control value.
In vitro assay for assessment of HIV antiviral activity
Antiviral efficacy against HIV may be tested as follows: Blood derived CD4+ T-lymphocytes and macrophages are isolated as described previously52. Briefly, human PBMCs were purified from fresh blood by banding on Ficoll–Hypaque (30 minutes, 800 g, 25 °C). Primary human CD4+ T cells were purified from PBMCs by positive selection with anti-CD4 Dynabeads and subsequent release using Detachabead. Cells were cultured in RPMI medium 1640 (Invitrogen) supplemented with 10 % FCS, MEM amino acids, l-glutamine, MEM vitamins, sodium pyruvate, and penicillin plus streptomycin and were subsequently activated with bacterial superantigen staphylococcal enterotoxin B (SEB; 100 ng/ml) and mitomycin C-killed PBMC from another donor (10:1 PBMC:CD4 cell ratio). Three days after stimulation, cells were split 1:2 in medium containing IL-2 (200 units/ml final concentration). Cultures were then split 1:2 every 2 days in IL-2 medium and infected with HIV at 7 days after stimulation. For generating primary human macrophages, monocytes were purified from human PBMCs by negative selection and activated and cultured at a cell concentration of 106/ml in DMEM, supplemented with 10 % FCS, MEM amino acids, l-glutamine, MEM vitamins, sodium pyruvate, and penicillin (100 units/ml), streptomycin (100 mg/ml), and 50 ng/ml recombinant human granulocyte–macrophage colony-stimulating factor (GM-CSF) and maintained at 37 °C in a humidified atmosphere supplemented with 5% CO2. To obtain monocyte-derived macrophages, cells were allowed to adhere to plastic and cultured for 6 days to allow differentiation.
CD4+ HeLa cells, Jurkat cells, activated CD4+ peripheral blood T-lymphocytes and macrophages (500,000 cells/100 µL) were incubated with pNL4.3-GFP (X4 virus) or pNL4.3-BaL-GFP (R5 virus) (100 ng of p24) in the presence of increasing concentrations of test article, 48 hours later, infection was scored by analyzing the percentage of GFP-positive cells by FACS and EC50 calculated.
SCID-hu Thy/Liv mouse model
Stocks of NL4-3 (X4 virus) were prepared by transfection of 293T cells and collection of supernatants on day 2. Supernatants were filtered, aliquoted and frozen at −80 °C until use. Amounts of virus were quantified by p24 ELISA and infectivity of viral stocks verified using CD4+ HeLa-betagalactosidase reporter cells.
Human fetal thymus and liver were obtained from Advanced Bioscience Resources in accordance with federal, state, and local regulations. Co-implantation of thymus and liver fragments under the kidney capsule to create SCID-hu Thy/Liv mice and inoculation of the Thy/Liv implants with HIV-1 (1000 TCID50 HIV-1 per Thy/Liv implant by direct injection) were carried out as described53,54. Male C.B-17 SCID (model #CB17SC-M, homozygous, C.B-Igh-1b/IcrTac-Prkdcscid) mice were obtained at 6–8 weeks of age from Taconic, and cohorts of 50–60 SCID-hu Thy/Liv mice were implanted with tissues from a single donor. Implants were inoculated 18 weeks after implantation with 50 µL of stock virus or complete DMEM (control infection) by direct injection. Animal protocols were approved by the TSRI Institutional Animal Care and Use Committee. Groups of 6 mice each were treated with NVP018 or alisporivir (ALV) at 50 or 100 mg/kg/day by twice-daily oral gavage until implant collection 2 weeks after inoculation.
Implants were dispersed through nylon mesh into single-cell suspensions and assessed for p24 by ELISA (p24 (ELISA): intracellular Gag pg/106 cells), for viral RNA (bDNA assay: copies/106 cells), and for depletion of thymocyte subsets by flow cytometry (% of CD3, CD4, CD8) as described previously29,53,55. Specifically, implant cells were stained with phycoerythrin cyanine dye CY7-conjugated anti-CD4 (BD Biosciences), phycoerythrin cyanine CY5.5-conjugated anti-CD8 (Caltag), allophycocyanin cyanine CY7-conjugated anti-CD3 (eBiosciences), and phycoerythrinconjugated anti-W6/32 (DakoCytomation). Cells were fixed and permeabilized with 1.2 % paraformaldehyde and 0.5 % Tween 20, stained with fluorescein isothiocyanate-conjugated anti-p24 (Beckman Coulter), and analyzed on an LSR II (BD Biosciences). After collecting 100,000 total cell events, percentages of marker-positive (CD4+, CD8+, and CD4+CD8+) thymocytes in the implant samples were determined by first gating on a live lymphoid cell population identified by forward- and side-scatter characteristics and then by CD3 expression. Total RNA was extracted from frozen thymocyte pellets using Trizol LS (Invitrogen) and resuspended in nuclease-free water. The capsid region of the HIV-1 gag was amplified by RT-PCR using 10 µL of purified RNA and AmpliTaq Gold (Applied Biosystems) according to the manufacturer’s instructions.
Supplementary Material
Significance.
Moss et al., describes the generation and preclinical analysis of a bacterial natural product with activity as a host targeted antiviral drug. This was generated by a combination of biosynthetic engineering and semisynthetic chemistry – bringing together the advantages of each method to more efficiently optimise a bacterially produced, genetically encoded natural product. The preclinical analysis reveals potent activity against HCV, HIV and HBV and a significantly improved profile as compared to both the parent natural product and the cyclosporine A (CsA) class of cyclophilin inhibitors, of which alisporivir is in phase III for treatment of chronic HCV infection.
Highlights.
Optimization and preclinical analysis of a bacterial natural product.
Combination of bioengineering and semisynthetic chemistry.
Preclinical analysis revealing potent antiviral activity.
Acknowledgements
We thank Drs. Wakita and Chisari for Huh-7.5.1 replicon cells, Shanghai Chempartner for chemical synthesis and ADMET studies and Cyprotex, UK and Solvo Biotechnology for ADME studies.
This work was supported by the U.S. Public Health Service grant no. AI087746 from the National Institute of Allergy and Infectious Diseases (NIAID).
NVP018 was formerly described as BC556 and is now referred to as NV556/OCB-030. It is being developed as an intravenous treatment by Neurovive Pharmaceutical AB as NVP019 and as an oral treatment for HBV by OnCore Biopharma, Inc. as OCB-030.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. 2007. [DOI] [PubMed] [Google Scholar]
- 2.Chu CK, Cutler HG. Natural products as antiviral agents. Plenum Press; 1992. [Google Scholar]
- 3.Hunt SR, Hernandez R, Brown DT. Role of the Vacuolar-ATPase in Sindbis Virus Infection. Journal of Virology. 2011;85:1257–1266. doi: 10.1128/JVI.01864-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc Natl Acad Sci U S A. 2002;99:3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Momose F, et al. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 6.Hutchinson CR. Drug synthesis by genetically engineered microorganisms. Biotechnology (N Y) 1994;12:375–380. doi: 10.1038/nbt0494-375. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson B, Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nature Chem Biol. 2007;3(7):379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 8.Gregory MA, Petkovic H, Lill RE, Moss SJ, Wilkinson B, Gaisser S, Leadlay PF, Sheridan RM. Mutasynthesis of rapamycin analogues through the manipulation of a gene governing starter unit biosynthesis. Angewandte Chemie (Int Ed.) 2005;44(30):4757–4760. doi: 10.1002/anie.200462784. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy J. Mutasynthesis, chemobiosynthesis, and back to semisynthesis: combining synthetic chemistry and bioengineering for diversifying natural products. Nat. Prod. Rep. 2008;25:25–34. doi: 10.1039/b707678a. [DOI] [PubMed] [Google Scholar]
- 10.Luvisetto S, Basso E, Petronilli V, Bernardi P, Forte M. Enhancement of anxiety, facilitation of avoidance behavior, and occurrence of adult-onset obesity in mice lacking mitochondrial cyclophilin D. Neuroscience. 2008;155(3):585–596. doi: 10.1016/j.neuroscience.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elrod JW, Wong R, Mishra S, Vagnozzi RJ, Sakthievel B, Goonasekera SA, Karch J, Gabel S, Farber J, Force T, Brown JH, Murphy E, Molkentin JD. Cyclophilin D controls mitochondrial pore-dependent Ca(2+) exchange, metabolic flexibility, and propensity for heart failure in mice. J Clin Invest. 2010;120(10):3680–3687. doi: 10.1172/JCI43171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colgan J, Asmal M, Luban J. Isolation, characterization and targeted disruption of mouse ppia: cyclophilin is not essential for mammalian cell viability. Genomics. 2000;68:167–178. doi: 10.1006/geno.2000.6295. [DOI] [PubMed] [Google Scholar]
- 14.Thali M, et al. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 15.Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 16.An P, et al. Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterji U, et al. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem. 2009;284(25):16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, et al. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporin resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathogens. 2009;5(8):e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damaso CR, Moussatche N. Inhibition of vaccinia virus replication by cyclosporin A analogues correlates with their affinity for cellular cyclophilins. J Gen Virol. 1998;79:339–346. doi: 10.1099/0022-1317-79-2-339. [DOI] [PubMed] [Google Scholar]
- 21.Qing M, Yang F, Zhang B, Zou G, Robida JM, Yuan Z, Tang H, Shi P-Y. Cyclosporin inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob Agents Chemother. 2009;53(8):3226–3235. doi: 10.1128/AAC.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ptak RG, et al. Inhibition of Human Immunodeficiency Virus Type 1 Replication in Human Cells by Debio-025, a Novel Cyclophilin Binding Agent. Antimicrob Agents Chemother. 2008;52(4):1302–1317. doi: 10.1128/AAC.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bienkowska-Haba M, Patel HD, Sapp M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009;5(7):e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki H, Mocarski ES, Kosugi I, Tsutsui Y. Cyclosporin inhibits mouse cytomegalovirus infection via a cyclophilin-dependent pathway specifically in neural stem/progenitor cells. J Virol. 2007;81(17):9013–9023. doi: 10.1128/JVI.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfefferle S, et al. The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kambara H, et al. Involvement of cyclophilin B in the replication of Japanese encephalitis virus. Virology. 2011;412:211–219. doi: 10.1016/j.virol.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, et al. Cyclosporin A Inhibits the Influenza Virus Replication through Cyclophilin A-Dependent and -Independent Pathways. PLoS ONE. 2012;7:e37277. doi: 10.1371/journal.pone.0037277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss SJ, et al. Sangamides, a new class of cyclophilin-inhibiting host-targeted antivirals for treatment of HCV infection. Med Chem Comm. 2012;3(8):944–949. [Google Scholar]
- 29.Stoddart CA, et al. Validation of the SCID-hu Thy / Liv Mouse Model with Four Classes of Licensed Antiretrovirals. PLoS ONE. 2007;8:1–11. doi: 10.1371/journal.pone.0000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu X, et al. Cloning, sequencing and characterization of the biosynthetic gene cluster of sanglifehrin A, a potent cyclophilin inhibitor. Mol Biosyst. 2010;7:852–861. doi: 10.1039/c0mb00234h. [DOI] [PubMed] [Google Scholar]
- 31.Kallen J, et al. Structure of Human Cyclophilin A in Complex with the Novel Immunosuppressant Sanglifehrin A at 1.6 Å Resolution. J Biol Chem. 2005;280(23):21965–21971. doi: 10.1074/jbc.M501623200. [DOI] [PubMed] [Google Scholar]
- 32.Sedrani R, et al. Sanglifehrin - Cyclophilin Interaction : Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data. J Am Chem Soc. 2003;125:3849–3859. doi: 10.1021/ja021327y. [DOI] [PubMed] [Google Scholar]
- 33.Kieser T, et al. Practical Streptomyces Genetics. John Innes Foundation; 1999. [Google Scholar]
- 34.Avila C, et al. Alisporivir (Deb025) treatment can be associated with transient, reversible and manageable hyperbilirubinemia, without signs of hepatoxicity: analysis of the Novartis clinical database. J Hepatol. 2012;56 s2:S428–S429. [Google Scholar]
- 35.Kapturczak MH, Kaplan B. Pharmacology of Calcineurin Antagonists. Transplant Proc. 2004;36:25S–32S. doi: 10.1016/j.transproceed.2004.01.018. 2004. [DOI] [PubMed] [Google Scholar]
- 36.Wring S, et al. In vitro models for assessing the relative risk of hyperbilirubinemia associated with cyclophilin inhibitor therapy. J Hepatol. 2010;52 s1:S263. [Google Scholar]
- 37.Gane EJ, et al. PSI-7977: ELECTRON Interferon is not required for Sustained Virologic Response in Treatment-Naive Patients with HCV GT2 or GT3; 62nd Annual Meeting of the American Association for the Study of Liver Disease (AASLD 2011); San Francisco. 2011. Abstract 34. [Google Scholar]
- 38.Sulkowski M, et al. Potent viral suppression with all-oral combination of daclatasvir (ns5a inhibitor) and GS-7977 (NS5B inhibitor), +/− ribavirin, in treatment-naive patients with chronic HCV GT1, 2, or 3. J Hepatol. 2012;56 s2:S560. [Google Scholar]
- 39.Pawlotsky J-M, et al. Alisporivir plus ribavirin is highly effective as interferon-free or interferon-add-on regimen in previously untreated HCV-G2 or G3 patients: SVR12 results from VITAL-1 Phase 2b study. J Hepatol. 2012;56 s2:S553. [Google Scholar]
- 40.Xia X-L, Shen Y, Zheng S-S. Inhibitory effect of cyclosporin A on hepatitis B virus replication in vitro and its possible mechanisms. Hepatobiliary Pancreat Dis Int. 2005;4(1):18–22. [PubMed] [Google Scholar]
- 41.Flisiak R, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 42.Kennedy J. Mutasynthesis, chemobiosynthesis, and back to semi-synthesis: combining synthetic chemistry and biosynthetic engineering for diversifying natural products. Nat Prod Rep. 2008;25:25–34. doi: 10.1039/b707678a. [DOI] [PubMed] [Google Scholar]
- 43.Eichner, et al. The Interplay between Mutasynthesis and Semisynthesis: Generation and Evaluation of an Ansamitocin Library. Angewandte Chemie (Int Ed.) 2012;51(3):752–757. doi: 10.1002/anie.201106249. [DOI] [PubMed] [Google Scholar]
- 44.Weist, et al. Fluorobalhimycin—A New Chapter in Glycopeptide Antibiotic Research. Angewandte Chemie (Int Ed.) 2002;41(18):3383–3385. doi: 10.1002/1521-3773(20020916)41:18<3383::AID-ANIE3383>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 45.Hojati, et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 2002;9:1175–1187. doi: 10.1016/s1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- 46.Goss, et al. An expeditious route to fluorinated rapamycin analogues by utilising mutasynthesis. Chembiochem. 2010;11(5):698–702. doi: 10.1002/cbic.200900723. [DOI] [PubMed] [Google Scholar]
- 47.Knobloch, et al. Unprecedented deoxygenation at C-7 of the ansamitocin core during mutasynthetic biotransformations Beilstein. J. Org. Chem. 2012;8:861–869. doi: 10.3762/bjoc.8.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almabruk, et al. Mutasynthesis of fluorinated pactamycin analogues and their antimalarial activity. Org Lett. 2013;15(7):1678–1681. doi: 10.1021/ol4004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obata Y, et al. Cyclophilin B in activation of interferon regulatory factor-3. J Biol Chem. 2005;280:18355–18360. doi: 10.1074/jbc.M501684200. [DOI] [PubMed] [Google Scholar]
- 50.Bobardt M, et al. HCV NS5A and IRF9 compete for CypA binding. Journal of Hepatology. 2013;58(1):16–23. doi: 10.1016/j.jhep.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katze MG, He Y, Gale MG. Viruses and interferon: a fight for supremacy. Nature Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 52.Bobardt M, et al. Hepatitis C virus NS5A anchor peptide disrupts human immunodeficiency virus. Proc. Natl. Acad. Sci. USA. 2008;105:5525–5530. doi: 10.1073/pnas.0801388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabin L, et al. Use of standardized SCID-hu Thy/Liv mouse model for preclinical efficacy testing of anti-human immunodeficiency virus type 1 compounds. Antimicrob Agents Chemother. 1996;40(3):755–762. doi: 10.1128/aac.40.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Namikawa R, et al. Long term human hematopoiesis in the SCID-hu mouse. J. Exp. Med. 1990;172(4):1055–1063. doi: 10.1084/jem.172.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoddart C, et al. Antiviral activity of 2′-deoxy-3′-oxa-4′-thiocytidine (BCH-10652) against lamivudine-resistant human immunodeficiency virus type 1 in SCID-hu Thy/Liv mice. Antimicrob Agents Chemother. 2000;44:783–786. doi: 10.1128/aac.44.3.783-786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.