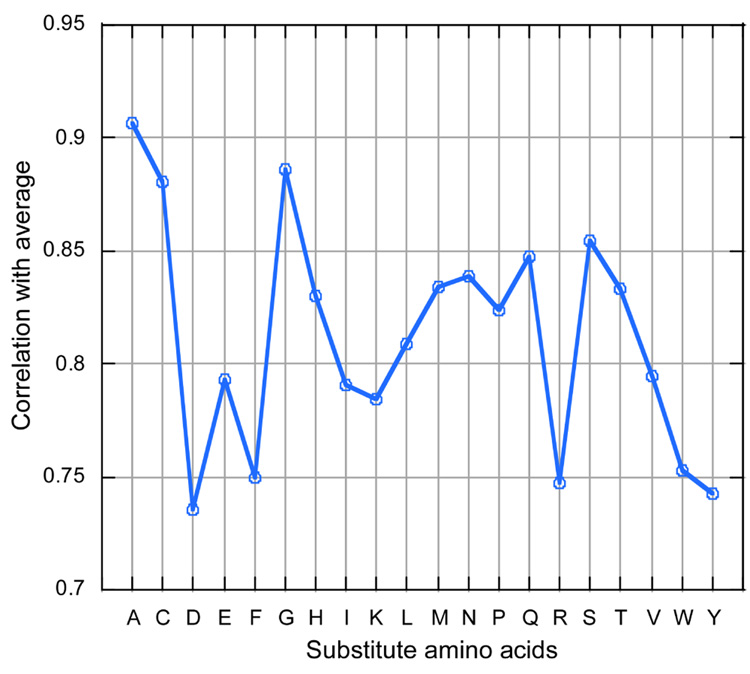
Figure 3. Average substitution effect correlated with single amino acid substitutions.
Among all single amino acid substitutions (at ASEdb mutant sequence positions), the distribution of predictions that best estimated the average was that of alanine, followed by cysteine, and glycine. These are also the amino acids that are often used in experimental mutagenesis studies to define functional sites.