Abstract
Accurate spatiotemporal assessment of extracellular vesicle (EV) delivery and cargo RNA translation requires specific and robust live-cell imaging technologies. Here we engineer optical reporters to label multiple EV populations for visualization and tracking of tumor EV release, uptake and exchange between cell populations both in culture and in vivo. Enhanced green fluorescence protein (EGFP) and tandem dimer Tomato (tdTomato) were fused at NH2-termini with a palmitoylation signal (PalmGFP, PalmtdTomato) for EV membrane labeling. To monitor EV-RNA cargo, transcripts encoding PalmtdTomato were tagged with MS2 RNA binding sequences and detected by co-expression of bacteriophage MS2 coat protein fused with EGFP. By multiplexing fluorescent and bioluminescent EV membrane reporters, we reveal the rapid dynamics of both EV uptake and translation of EV-delivered cargo mRNAs in cancer cells that occurred within one-hour post-horizontal transfer between cells. These studies confirm that EV-mediated communication is dynamic and multidirectional between cells with delivery of functional mRNA.
Cells release exosomes, microvesicles and apoptotic blebs as a conduit for intercellular communication without direct cell-to-cell contacts1–4. These vesicles, collectively termed extracellular vesicles (EVs), are capable of horizontal transfer of lipids, (glyco)proteins and nucleic acids from EV-donor cells to neighboring and/or distal recipient cells5,6. Previous reports support the functionality of EV-delivered mRNAs7,8, but direct visualization of RNA cargo and mRNA fate has been lacking, essential in understanding the kinetics and spatiotemporal arrangements. While different methods including electron microscopy and atomic force microscopy can be used to study the physical properties of EVs, live-cell imaging at high spatial resolution generally requires fluorescent microscopy techniques using EV-associated proteins fused to fluorescent proteins, and/or fluorescent chemical labeling6,9–12.
Current EV labeling methods have some limitations. For fluorescent proteins conjugated to proteins enriched in EVs, labeling may be restricted to subpopulations of EVs, limiting their wider use to observe multiple EV types. On the other hand, fluorescent dyes for EV lipid labeling including PKH may not reflect true half-life of EVs and can be retained in association with other lipid entities for long periods, thus misguiding spatiotemporal assessment of EV dynamics, especially over extended periods13. In addition, studying the fate of mRNAs transcribed and packaged into EVs released by cells has been challenging due to their low quantity14, thus demanding a highly sensitive imaging technique. Furthermore, limitations on EV reporter choice render current studies on EV-mediated communication mainly to unidirectional EV transfer from one cell type to another2, whereas EV exchange under physiological and/or pathological condition is expected to be multidirectional between cell populations.
S-Palmitoylation through a thioester linkage between the sulfhydryl group on cysteines and the fatty acid palmitic acid enables association of proteins with cellular membranes15. By fusing fluorescent proteins with a consensus palmitoylation sequence, these reporters can be transferred to cellular membrane, enabling whole cell labeling16,17. Since EVs are derived from the plasma membrane either via inward invagination of endosomes to form multivesicular bodies releasing exosomes18 or outward budding and shedding for microvesicles and apoptotic blebs released from the cell surface19–21, we hypothesized that tagging the plasma membrane with fluorescent proteins would enable labeling of multiple EV types.
Bioluminescence reporters have also been widely adapted to study biological processes including gene expression based on their sensitivity and low background signal without the need of an excitation source to emit light22. Specifically, Gaussia luciferase (Gluc), which is over 1,000-fold brighter than commonly used luciferases23 catalyzes flash bioluminescence with rapid signal decay, making it an ideal reporter to study temporal properties of minute events, such as translation of EV-delivered mRNAs.
In this study, we develop a fluorescent EV labeling strategy to achieve live-cell imaging of EV release, uptake, and exchange between different cell populations, as well as microscopic quantification and flow cytometry analysis. By multiplexing with the bacteriophage MS2 coat protein (MS2CP) mRNA fluorescent reporter system24 and a bioluminescent EV reporter13, we visualize EV-mRNA cargo and further reveal the temporal dynamics of EV uptake and translation of EV-delivered mRNAs.
RESULTS
PalmGFP and PalmtdTomato label EVs
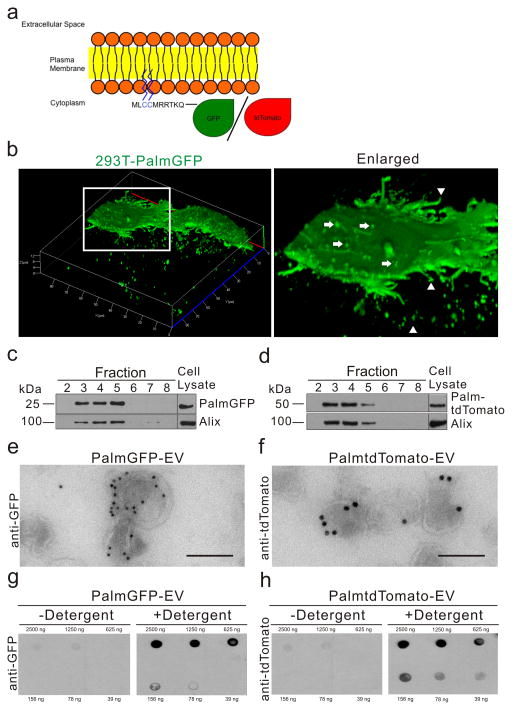
To generate fluorescent EV reporters, a palmitoylation signal was genetically fused in-frame to the N-terminus of EGFP (PalmGFP)16,17 or tdTomato25 (PalmtdTomato) (Fig. 1a). 293T cells were then stably transduced with a lentivirus vector encoding either PalmGFP (293T-PalmGFP) or PalmtdTomato (293T-PalmtdTomato) to examine EV reporter expression. Live-cell confocal microscopy of 293T-PalmGFP cells showed that PalmGFP uniformly labels the plasma membrane and reveals budlike structure on their surface and processes (Fig. 1b). PalmGFP+ EVs of varying sizes were also detected around 293T-PalmGFP cells, suggesting the reporter labels multiple EV subtypes (Fig. 1b). To verify specificity of the EV reporters, EVs isolated from 293T-PalmGFP or 293T-PalmtdTomato cells were isolated by sucrose density gradient centrifugation, and PalmGFP and PalmtdTomato labels were found specifically in fractions 3–5, which are also the fractions exhibiting the exosomal/microvesicle marker Alix26,27 (Fig. 1c, d; Supplementary Fig. 1). To further examine if PalmGFP or PalmtdTomato labels EVs in association with membranes, transmission electron microscopy was applied in combination with immunogold labeling of PalmGFP or PalmtdTomato. These experiments showed that EVs were indeed labeled on their membranes, and not within the EVs (Fig. 1e, f, Supplementary Fig. 2). To characterize the orientation of PalmGFP and PalmtdTomato on the EV membranes, labeled EVs were dot blotted which showed predominant PalmGFP/tdTomato signals associated with the inner membrane (Fig. 1g, h). Under this experimental setup, these fusion proteins should only be readily detected in a concentration-dependent manner in the presence of a detergent (Tween-20), which opens EV membranes to allow antibody entry and labeling of the reporter proteins. In fact, only blots immunolabeled in the presence of the detergent revealed PalmGFP and PalmtdTomato signals when compared to the control (without detergent).
Figure 1. Palmitoylated-GFP and -tdTomato are trafficked to the plasma membrane and EVs.
(a) Schematic diagram of EV membrane labeling with palmitoylated GFP (PalmGFP) or tdTomato (PalmtdTomato). (b) Live-cell confocal microscopy of stable PalmGFP-expressing 293T cells releasing EVs. Left panel: 3D reconstruction of confocal Z-stack images demonstrating EV release from stable 293T-PalmGFP cells into surrounding regions. Right panel: Enlarged image of boxed region, showing budlike structure from the surface (arrows) as well as processes extending from cells (arrow heads). (c, d) Western blot analysis of protein extracted from fractions following sucrose gradient centrifugation of isolated EVs. PalmGFP (c) and PalmtdTomato (d) were detected in fractions exhibiting the EV marker, Alix (95 kDa). (e, f) Transmission electron micrograph (TEM) showing PalmGFP and PalmtdTomato labeling of EVs on the membrane following immunolabeling with anti-GFP (e) or anti-tdTomato (f) and secondary gold-conjugated secondary antibodies. Bar, 100 nm. (g) Demonstration that PalmGFP and PalmtdTomato labels the inner membrane of EVs. PalmGFP- or PalmtdTomato EVs were dot blotted on nitrocellulose membrane in a dose range followed by immunoprobing with anti-GFP or anti-tdTomato, respectively, and horseradish peroxidase (HRP)-conjugated secondary antibodies in the presence or absence of detergent [0.1% (v/v) Tween20] for chemiluminescence detection.
PalmGFP/tdTomato label different sized EVs
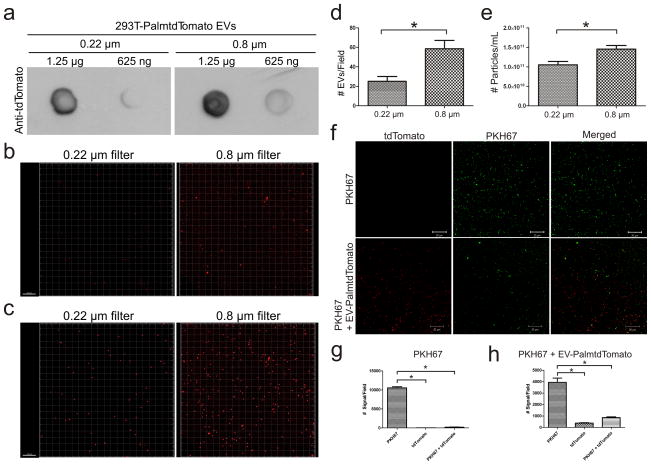
To test if the palmitoylated fluorescent EV reporters label more than one population of EVs, conditioned medium from 293T-PalmtdTomato cells was filtered through either a 0.22 or 0.8 μm filter prior to EV isolation. PalmtdTomato was readily detected in EVs isolated from both 0.22 and 0.8 μm filtered samples, indicating the reporter labels different sized EVs (Fig. 2a). Given the conventional dogma that exosomes range in size from 40 to 100 nm, while microvesicles falls between 100 and 1,000 nm in diameter28, the number of isolated EVs in the 0.22 μm filtered sample should be less than that in the 0.8 μm filtered sample. In fact, we did observe fewer EVs per field in the 0.22 μm sample as compared to the 0.8 μm sample, as assessed by semi-quantitative microscopic and quantitative nanoparticle tracking analysis (NTA) of vesicle number (Fig. 2b–e).
Figure 2. PalmtdTomato labels different sized EVs and co-labels with PKH.
(a) Dot blot detection of PalmtdTomato+ EV isolated from 0.22 or 0.8 μm filtered conditioned medium of 293T-PalmtdTomato cells. (b) Confocal microscopy of EVs isolated from 0.22 or 0.8 μm filtered conditioned medium of 293T-PalmtdTomato cells. Bar, 10 μm. (c) Digital display of PalmtdTomato+ EVs signals as spheres. Bar, 10 μm. (d) Quantification of PalmtdTomato+ EV signals per field in 0.22 or 0.8 μm filtered samples. *p<0.001 by two-tailed, t-test with 14 replicates. (e) Nanoparticle tracking analysis (NTA) of PalmtdTomato+ EVs isolated from 0.22 and 0.8 μm filtered samples. *p<0.05 by two-tailed t-test with 6 replicates. The results are presented as the mean ± s.e.m. (f) EV-depleted medium (top rows) or EVs isolated from 293T-PalmtdTomato cells (bottom rows) were stained with PKH67 followed by a 1 h wash in PBS at 100,000 g. Bar, 20 μm. (g, h) Semi-quantification of EV signals from PKH67-stained EV-depleted medium (g), or PalmtdTomato+ EVs co-labeling with PKH67 (h). *p<0.05 by repeated measures analysis of variance (ANOVA) followed by Tukey’s post hoc test with 4 replicates. The results are presented as the mean ± s.e.m.
PalmtdTomato labels EVs more specifically than PKH67 dye
PKH fluorescent dyes, which label cell membranes by the insertion of their aliphatic chains into the lipid bilayer, have been widely used to label EVs based on their intense signal and long half-life6,7,29–32. However, we recently reported that the half-life of PKH dyes likely outlast that of EVs in vivo, thereby yielding inaccurate spatiotemporal assessments of EV fate13. We examined whether EV labeling with PKH dyes is entirely EV-specific and found that PKH67-labeled cell-free culture medium, initially used as a negative control, produced a higher signal than PKH67-labeled EVs isolated from 293T-PalmtdTomato cells under the same conditions (Fig. 2f–h). These results indicate that PKH67 not only labels EVs but may also aggregate and/or form micelles33, some of which are associated with pelleted EVs following an ultracentrifugation-based isolation procedure, resulting in false-positive signals for EVs. Although not statistically significant, we noted a small fraction of PKH67-labeled PalmtdTomato+ EV that are not detected with PKH67 alone (Fig. 2h), suggesting that it is PalmtdTomato protein that co-purified with EVs and/or there can be an artifact of Brownian motion which produces a spatial shift between the tdTomato and PKH67 channels during image acquisition, thereby yielding non-PKH67 labeled PalmtdTomato+ signals.
EV exchange between cell populations
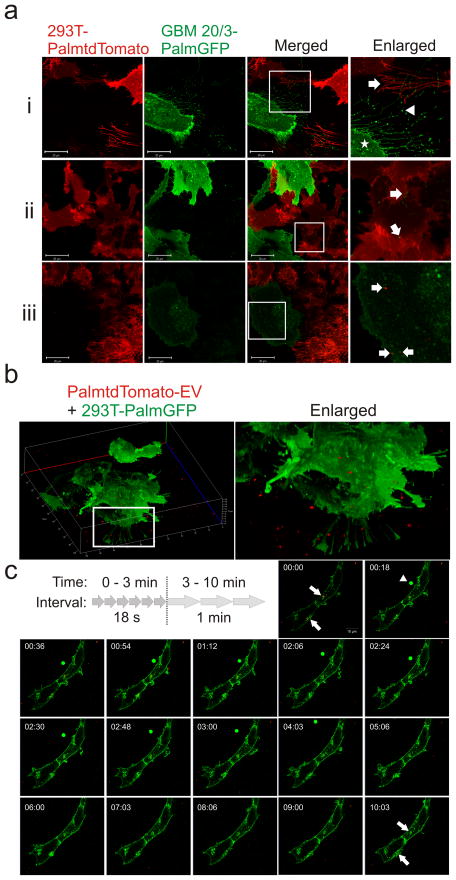
It is widely believed that EVs are dynamically exchanged between cell populations. However, most studies to-date, possibly due to a lack of a dual imaging system, have primarily focused on one-way delivery of EVs to cells. To demonstrate bi-directional EV exchange between cells without an intermediate purification step, we co-cultured 293T-PalmtdTomato and primary glioblastoma cells expressing PalmGFP (GBM-PalmGFP), and observed EV exchange between the differentially-labeled cell types (Fig. 3a). Both cell types were also found to extend fine, long projections that can measure up to 60 μm in length and 200 nm in diameter, reminiscent of nanotubes34. Interestingly, some of the projections appeared to display EV-like structures at their tips.
Figure 3. Live-cell imaging of EV exchange and uptake.
(a) Stably transduced 293T-PalmtdTomato and primary GBM-PalmGFP cells were co-cultured and imaged with confocal microscopy for cellular interactions and EV exchanges between the cells. Magnification of boxes in merged panels are shown in far right panels. (i) Multiple thin cellular projections were readily observed extending from both GBM (arrow) and 293T (arrowhead) cells. Notably, many GBM cellular projections exhibited vesicle-like structure at the tips of the projections. Vesicle-like structures were also detected on the surface of GBM cells (star). (ii) PalmGFP+ EVs from GBM cells are found inside of 293T-PalmtdTomato cells (arrows). (iii) PalmtdTomato+ EVs from 293T cells are detected inside GBM-PalmGFP cells (arrows). Bar, 20 μm. (b) 3D reconstruction of Z-stack images showing 293T-PalmGFP cells exposed to PalmtdTomato+ EVs for 1.5 h. (c) Time-lapsed confocal imaging of PalmtdTomato+ EV attachment on 293T-PalmGFP cells at 2.45 h. Images shown at an 18 sec interval from 0 to 3 min followed by a 1 min interval from 3 to 10 min. PalmtdTomato+ EVs were surrounded by 293T-PalmGFP cell membranes (arrows) within a minute, and the pattern lasted until the end of the experiment (10 min). Notably, a round-shaped, PalmGFP-labeled entity with a diameter ≥ 4 μm (arrowhead) in the extracellular space was also observed from 00:18 to 04:03 min. Bar, 10 μm.
High-resolution tracking of EV uptake
To examine if the fluorescent reporters can be used to track EVs at high-resolution, EVs isolated from 293T-PalmtdTomato cells were seeded onto 293T-PalmGFP recipient cells. Using live-cell confocal microscopy, PalmtdTomato+ EVs were detected around or on the 293T-PalmGFP cells (Fig. 3b), demonstrating that these reporters can also be used to distinguish recipient cells from the EVs. Further, time-lapse imaging of PalmtdTomato+ EV exposed cells showed that EV attachment and uptake by recipient cells could be tracked in real-time using these reporters (Fig. 3c, Supplementary Movie 1, Supplementary Fig. 3). A PalmGFP vesicle-like signal with an estimated diameter of 4 μm near the recipient cells starting at 0:18 sec was also observed. Given its relatively large size, it may represent cell debris and/or a type of oncosomes as previously described35, demonstrating that the fluorescent EV reporters can also label larger EVs. To further validate palmitoylated fluorescent EV uptake by the recipient cells, we demonstrated PalmtdTomato+ EV uptake by Gli36-PalmGFP glioma cells by fluorescence activated cell sorting (FACS) analysis in the presence or absence of heparin, which reduced EV uptake as previously reported36 (Supplementary Fig. 4).
Simultaneous visualization of EV and EV-packaged mRNA
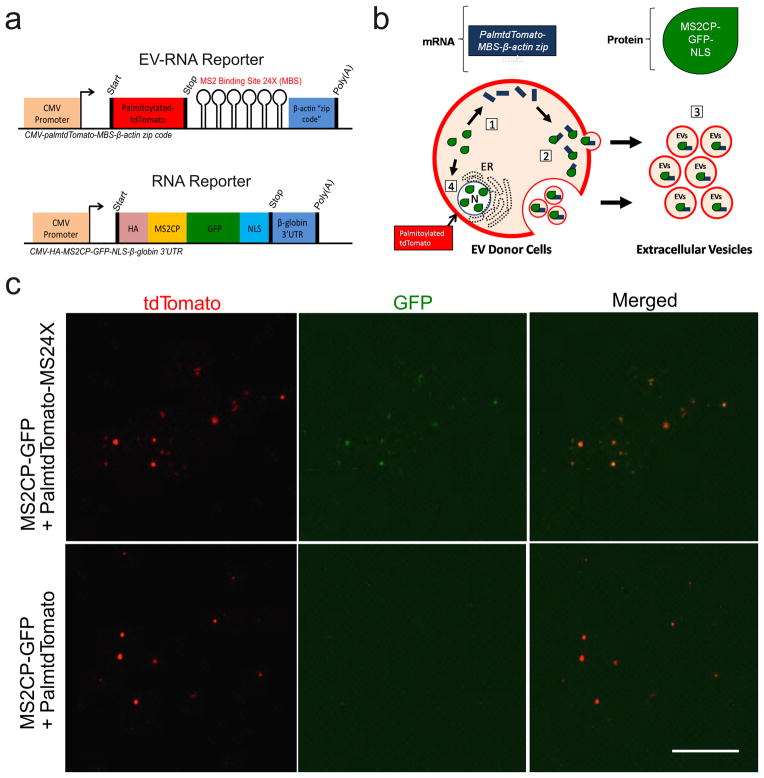
Next, we designed a dual-function reporter for simultaneous visualization of both EV membranes and EV-RNAs (Fig. 4a). In this reporter scheme, PalmtdTomato protein was used for EV visualization while its transcript was tagged with a repeated MS2 RNA binding sequence in the 3′ UTR (PalmtdTomato-MS24X) for EV-RNA detection by bacteriophage MS2 coat protein fused with EGFP (MS2CP-GFP) expressed in the same cells (Fig. 4b). EVs isolated from Gli36 glioma cells co-expressing PalmtdTomato-MS24X and MS2CP-GFP exhibited clear co-localization between PalmtdTomato (EV) and MS2CP-GFP signals (PalmtdTomato RNA:MS2CP-GFP complex), whereas only PalmtdTomato (EV) but no MS2CP-GFP was detected in EVs from Gli36 expressing PalmtdTomato lacking the MS24X tag along with MS2CP-GFP (negative control; Fig. 4c). To further confirm that the co-localization of signals between PalmtdTomato and MS2CP-GFP is not a result of unbound MS2CP-GFP aggregates co-purified with PalmtdTomato+ EVs, the EV samples were imaged in the presence or absence of trypsin to remove EV surface bound proteins, and the co-localization was unaffected (Supplementary Fig. 5). To assess the diffraction limit of the microscope, we transfected siGLO RISC-free siRNA into 293T-PalmGFP cells and observed transfer of siRNA-packaged EVs to the Gli36 GBM recipient cells with an estimated diffraction limit of 200 – 300 nm (Supplementary Fig. 6).
Figure 4. Visualization of EVs and packaged mRNAs.
(a) Dual-function EV-RNA reporter system enabling EV and EV-RNA labeling; two constructs encoding either PalmtdTomato protein with RNA reporter transcripts containing repeat MS2 RNA binding sequences (MS24X; PalmtdTomato-MS24X; top), or MS2-GFP coat protein (MS2CP) with a nuclear localization signal (NLS) (bottom). (b) Diagram of vesicle RNA labeling process: [1] RNA reporter (blue bar) allows detection of EV-RNA reporter transcripts through binding of MS2 coat protein fused with GFP (MS2CP-GFP-NLS, green dewdrop) to PalmtdTomato-MS24X sequence in mRNA; [2] PalmtdTomato-MS24X:MS2CP-GFP RNA reporter complexes are packaged into EVs; [3] EVs are released from cells by budding from the cell membrane and/or fusion of multivesicular bodies with the cell membrane; and [4] Nonspecific EV packaging of MS2CP-GFP-NLS is minimized by translocating the unbound RNA MS2CP-GFP reporter into the nucleus via the NLS signal. Abbreviations: CMV, cytomegalovirus enhancer/promoter; HA, hemagglutinin tag; NLS, nuclear localization signal; UTR, untranslated region. (c) EVs were isolated from human Gli36 glioma cells expressing MS2CP-GFP with either PalmtdTomato-MS24X or PalmtdTomato alone (negative control for MS24X mRNA binding sites). Confocal microscopy imaging showed that only EVs isolated from cells co-expressing MS2CP-GFP and PalmtdTomato-MS24X exhibited co-localization between EVs (tdTomato) and MS2CP-GFP:MS24X mRNA complexes (GFP; top row). The negative control for MS24X (PalmtdTomato) showed only membrane labeled EVs (tdTomato). Bar, 10 μm.
Visualizing tumor-derived EVs in vivo
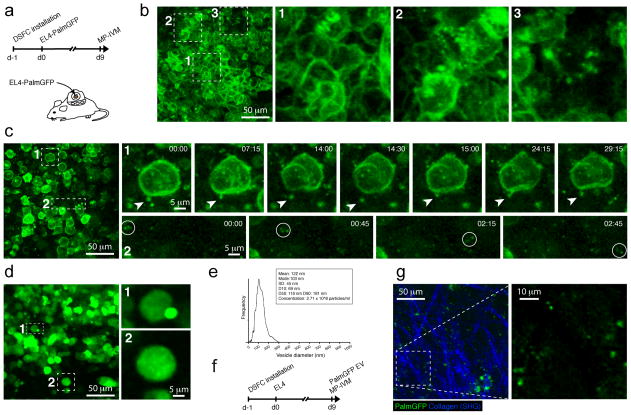
EV release and uptake are considered as potential mechanisms by which tumor cells communicate with each other and with their environment31. To test the general feasibility of visualizing EVs in tumor tissue by multiphoton intravital microscopy (MP-IVM), we stably expressed PalmGFP in the mouse thymoma cell line EL4 (EL4-PalmGFP) and implanted cells in dorsal skinfold chambers (DSFCs), where they formed solid tumors as described37,38 (Figure 5a). When we imaged these tumors by MP-IVM 9 days later, we observed green fluorescent punctae that were either intracellular, associated with cytoplasmic membranes, or outside of EL4-PalmGFP tumor cells. The density of fluorescent punctae, which based on their origin likely represent different classes of tumor-derived vesicles, varied across the tumor environment. The lowest densities were typically observed in areas of highest tumor cell density at the core of the tumor parenchyma (Fig. 5b, Supplementary Movie 2). More peripheral regions, in contrast, where the tumor parenchyma interfaces with the tumor stroma and where the density of tumor-infiltrating cells is lower while tumor-infiltrating immune cells, such as T cells and myeloid antigen-presenting cells, are more abundant39 showed the greatest density of fluorescent vesicles (Figure 5c, Supplementary Movie 3). Time-lapse recordings showed that some of the larger EVs (estimated 1 μm diameter) often remained tethered to tumor cells for prolonged periods of time (Figure 5c, subpanel 1, and Supplementary Movie 4). We did not observe this behavior for smaller EVs, which may be due to the enhanced difficulty of tracking these by imaging of thin optical sections in the three-dimensional environment of tumors in vivo (as opposed to the two-dimensional environment of a culture dish). However, both small and larger EVs were observed to traffic individually within the tissue or to group in small, fast-moving clusters, suggesting that they were attached to or had been internalized into a motile, non-visualized cell population, such as tumor-infiltrating immune cells (Figure 5c, subpanel 2, and Supplementary Movie 5).
Figure 5. In vivo visualization of EVs.
(a) PalmGFP-expressing EL4 tumors were implanted in pre-installed dorsal skinfold chambers (DSFCs) in C57BL/6 mice, and imaged 9 days later by MP-IVM. (b) Intravital micrograph from the central region of an EL4 tumor with regions characterized by the absence (subpanel 1), low (subpanel 2) and intermediate density (subpanel 3) of detectable PalmGFP+ vesicles. (c) Intravital micrograph of a peripheral region of a tumor with a high density of extracellular PalmGFP+ vesicles. Time-lapse recordings highlighting a tethered vesicle (subpanel 1) and a motile cluster of three vesicles (subpanel 2). Time is shown as min:sec. (d) An identical experiment was performed as described for (a-c), but EL4 tumor cells expressing soluble GFP instead of PalmGFP were used instead. Note the absence of small micron- and submicron-sized particles (e) Nanoparticle tracking analysis of vesicles purified from the supernatant of PalmGFP-expressing EL4 tumor cells. (f) Non-fluorescent EL4 tumor were implanted into DSFCs and purified PalmGFP+ vesicles directly injected into the tumor on day 9. (g) Intravital micrograph of an EL4 tumor 60 min after injection of GFP+ EVs.
EL-4 tumors expressing soluble instead of membrane-targeted GFP also produced a small number of fluorescent objects of subcellular size in vivo. However, these objects, which likely represent tumor cell apoptotic bodies, were much larger (up to 5 μm in diameter) and less numerous than the small membrane-derived fluorescent punctae observed in EL4-Palm-GFP tumors (Figure 5d, Supplemental Movie 6 and 7), indicating that the latter are indeed tumor membrane-derived vesicles, and not simply tumor cell debris that has been ingested by non-visualized phagocytes.
To assess whether the EVs detectable in EL4-PalmGFP tumors are likely to correspond to EVs as characterized in vitro, we purified EVs of sizes up to 300 nm from these tumor cells in vitro (Figure 5e), and injected these into established, non-fluorescent EL4 tumors in DSFCs. One hour later we observed, amidst the anatomical landmark structures of tumoral collagen fibers, GFP+ EVs with a similar size distribution as observed in EL4-PalmGFP tumors (Figure 5f). While our imaging system is likely not sensitive enough to detect the smallest EVs, this suggests that EVs rapidly coalesce in tumor tissue, probably in part through uptake by non-tumor cells. Collectively, these observations establish the feasibility of investigating the dynamic behavior of tumor-derived EVs in vivo through fluorescent protein-labeling of tumor cell membranes.
EV uptake and nascent EV-RNA translation
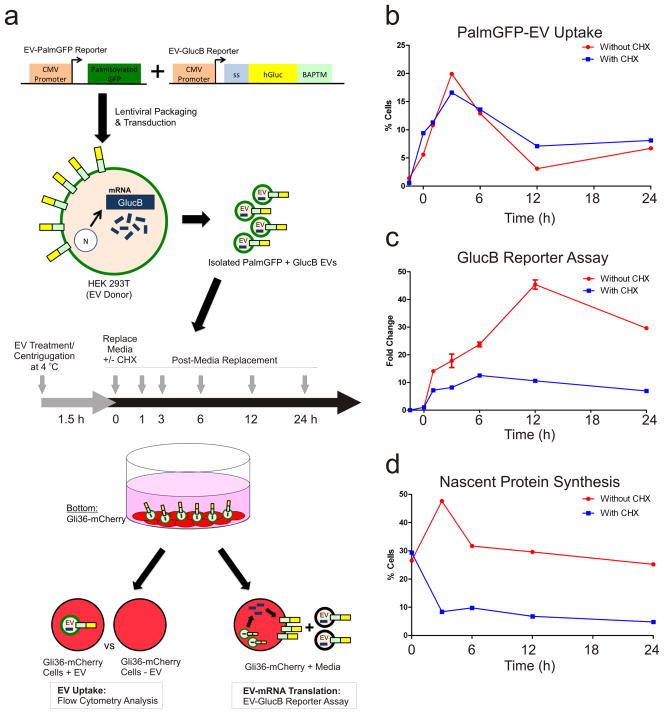
While EVs have been shown to mediate RNA and protein transfer to elicit phenotypic changes in recipient cells5, detailed temporal properties of these events are still under study. We multiplexed the fluorescent (PalmtdTomato) and bioluminescent (EV-GlucB)13 EV membrane reporters to monitor time of EV-uptake and EV-mRNA translation in the recipient cells (Fig. 6a). Using centrifugation at 4 °C to promote EV uptake and restrict the time window of uptake followed by FACS analysis, we determined that EVs from 293T-PalmGFP+GlucB cells were actively taken up by the recipient Gli36-mCherry glioma cells, between 0 to 3 h following EV treatment (Fig. 6b; Supplementary Fig. 7). This upward trend was followed by a decrease in EV signal from 3 to 12 h, suggesting that EVs and associated proteins are processed and degraded during this time. Interestingly, the signal came to a plateau from 12 to 24 h, but did not reach the baseline, implying that: i) a small proportion of EVs may remain intact in the recipient cells; and/or ii) the EVs were not fully degraded and some EV membrane (with the PalmGFP reporter) remained in the cells. Treatment with the protein translation inhibitor, cycloheximide (CHX) did not appear to affect EV uptake and dynamics in the recipient cells (Fig. 6b).
Figure 6. EV uptake and EV-RNA Translation.
(a) Schematic of two expression reporters used to detect EV uptake (PalmGFP) and translation of EV-delivered GlucB mRNA (luciferase activity). 293T cells (donors) were stably transduced with lentivirus vectors expressing these reporters. EVs were isolated from conditioned medium of these cells and added to Gli36-mCherry glioma cells followed by centrifugation at 4 °C to facilitate EV docking on cells while minimizing EV uptake. PalmGFP+GlucB+ EV-containing media was then replaced with EV-depleted media in the presence and absence of cycloheximide (CHX) and cells were incubated at 37 °C for 24 h. Medium and cell samples were collected at indicated time points to assess EV uptake (fluorescence and luciferase activity) and EV-mRNA translation (luciferase activity). Abbreviations: CMV, cytomegalovirus enhancer/promoter; ss, signal sequence; BAPTM; biotin acceptor peptide with transmembrane domain of platelet-derived growth factor receptor. (b) Flow cytometry analysis of the percentage of Gli36-mCherry cells positive for PalmGFP+ EV uptake in the presence or absence of CHX. Cells that have taken up EVs exhibited signal for both PalmGFP (EVs) and mCherry (recipient cells), whereas cells without or with low levels of EVs showed only mCherry (see also Supplementary Fig. 5). (c) GlucB reporter assay was performed to detect translation of EV-delivered GlucB mRNA by Gli36-mCherry cells expressed as fold change. (d) Flow cytometry analysis of nascent protein synthesis in Gli36-mCherry cells revealed by Click-iT® HPG labeling and Alexa Fluor®488 conjugation expressed as percentage of cells positive for Alexa Fluor® 488 (see also Supplementary Fig. 6).
To detect and monitor translation of EV-delivered mRNA in parallel with EV uptake, EV-treated cells and conditioned media were collected and luciferase activity of the GlucB reporter was measured. If the recipient cells translate EV-delivered GlucB mRNA, an increase in GlucB signal should be observed, whereas treatment with CHX should prevent GlucB mRNA translation and reduce any increase in the signal. Remarkably, the elevation in GlucB signal was observed as early as 1 h post-EV exposure, indicating that translation of EV-delivered GlucB mRNA began soon after EV uptake by the cells (Fig. 6c). The increase continued and reached a peak at 12 h and was followed by a decline between 12 and 24 h. This pattern directly correlated with levels of EV-uptake (Fig. 6b) where highest and lowest percentage of EV-containing cells was observed between 0 and 12 hr, and 12 to 24 h, respectively. In addition, samples treated with CHX consistently showed a significantly lower level of GlucB signal when compared to non-CHX-treated samples, with these translation-blocked samples serving as a baseline reading for GlucB reporter protein directly transferred by the EVs. Furthermore, nascent protein synthesis was confirmed to be suppressed by CHX treatment throughout the experiment as revealed by Click-iT® homopropargylglycine (HPG) labeling and FACS analysis (Fig. 6d; Supplementary Fig. 8). We thereby successfully monitored and revealed temporal dynamics of both EV uptake and translation of EV-delivered mRNA.
Next we wanted to further confirm that RNA transfer by EVs could indeed induce a physiological response in recipient cells. Given the pivotal role of the Nuclear Factor Kappa B (NFκB) in different physiological processes and the fact that its regulation can be modulated through various exogenous factors40, we investigated NFkB activity following EV-mediated RNA transfer. 293T recipient cells were engineered to stably express a bioluminescent NFkB reporter driving the expression of Gluc (NFkB-Gluc)41,42 and treated with 293T-EVs. We observed a significant elevation in NFkB activity from 24 to 72 h followed by a decline at 96 h post-EV treatment (Supplementary Fig. 9a, b). Since EVs contain lipids, proteins and other nucleic acids in addition to RNA, the observed increase in NFkB activity could be attributed to multiple factors. To distinguish whether this effect on NFkB is, at least in part, contributed by EV-delivered RNA, the recipient cells were transfected with varying amount of RNA isolated from the EVs and showed a dose-dependent rise in NFkB activity from 24 to 72 h post-EV-RNA transfection, corroborating the trend of EV-induced NFkB activation (Supplementary Fig. 9c). While it remains to be determined which RNAs resulted in the current observation, we hereby demonstrated that the delivery of EV-RNA alone could trigger cell signaling and induce an NFkB response over an extended period of time in recipient cells.
DISCUSSION
Given that most EVs are nanosized as opposed to micrometer-sized cells, an ideal EV labeling method for imaging at subcellular resolution should meet the following criteria: i) specific labeling of EVs; ii) stable and sufficient signal to detect nanoscaled EVs from background noise; and iii) a half-life consistent with that of EVs. The palmitoylated (Palm) fluorescent EV reporters described here achieved these needs for EV visualization, enabling: (a) labeling of multiple EV types irrespective of their biogenesis; (b) semi-quantification of EVs; (c) time-lapse live-cell imaging of EV release and uptake; (d) EV exchange between different cell populations; (e) intravital EV imaging; and (f) FACS analysis of EV uptake.
PalmGFP and PalmtdTomato, in contrast to labeling with EV-enriched protein conjugated fluorescent proteins such as CD63-GFP43, are designed to be a general EV labeling strategy to visualize and track multiple EV subtypes. This was shown by detection of PalmGFP+ and PalmtdTomato+ EVs in both 0.22 and 0.8 μm sized EV populations, as well as sucrose density gradient with EV-marker proteins, such as Alix. Interestingly, using these reporters, we also observed EV-like structures at the tips of cell projections. As nanofilaments were recently found to be present on glioblastoma exosomes using peak force microscopy44, these cell projections with EV-like structures may represent another mechanism by which EVs are released in addition to the previously described plasma membrane budding of microvesicles and exosome release from the multivesicular bodies. By labeling different cell populations in co-culture, it was also possible to monitor the exchange of EVs between cell types, demonstrating that EV transfer is dynamic and bidirectional.
PKH, along with other lipophilic dyes such as DiI, represent one of the most commonly used dyes to label EVs6,7,32. However, here we showed that PKH67 staining of EVs is not EV-specific, and that it labels other lipid-containing entities in the extracellular space, and presumably in recipient cells, potentially leading to false-positive signals. Furthermore, PKH dyes were reported to have an in vivo half-life of 5 to >100 days45. While its long half-life may be advantageous for some purposes, the dye may outlast labeled EVs following EV processing/degradation by cells, leading to misinterpretation of dye residues and/or recycled dye-labeled membrane as originally labeled EVs. In fact, our recent study showed that intravenously-administered EVs have an in vivo half-life of less than 30 min in most tissues13. Using PalmGFP+ EVs, we showed that the in vitro half-life of EVs in recipient cells is shorter than 24 h, which implies that any vesicle-like structures labeled with PKH dyes after this time are unlikely to be EVs. In addition, since EV surface proteins, at least in part, have been reported to be responsible for the binding and subsequent uptake of EVs by the recipient cells46,47, it is ideal to minimize perturbation to the EV surface proteome during labeling. Here we demonstrated that both PalmGFP and PalmtdTomato predominantly label the inner EV membrane, thereby reducing potential disturbance to the EV surface protein composition.
We also confirm here, using direct in vivo visualization through MP-IVM that tumor cells release EVs of varying sizes, at least some of which are likely taken up by migratory, non-tumor cells, likely migratory immune cells. Simultaneous visualization of different immune cell subsets in future studies will provide clues to reveal the identity of the cell types actively taking up EVs released by tumor cells. Implantation of mixtures of tumor cell clones expressing fluorescent proteins in complementary colors will also allow us to monitor EV exchange between tumor cells, which may be masked in our current system. Interestingly, EV density was highest at the tumor-parenchyma border, suggesting that EV-formation or release may be enhanced as a result of the interaction of malignant cells with the surrounding normal tissue microenvironment, including the immune cells.
We witnessed both endogenously generated and exogenous RNA cargos of EVs using the MS24X RNA sequence in combination with MS2CP-GFP in the donor cells48 or by transfection of donor cells with fluorescently labeled siRNA, respectively. We further investigated nascent translation of EV RNA cargo and revealed translation of EV-mRNA within 1 h following EV uptake by recipient cells. This timeframe coincides with that of liposome-delivered mRNAs where translation occurs within 1 h post-transfection49. Interestingly, the decrease in GlucB signal from 12 to 24 h corresponds to a reduction in the percentage of cells showing labeled EVs present during this same time period, suggesting that the decreased GlucB signal is a result of decreasing availability of external EVs combined with internal degradation of GlucB transcripts.
EV uptake has been shown to occur via multiple routes, including a direct fusion between EVs and the plasma membrane50, as well as EV internalization via lipid raft-, clathrin- and calveolae-dependent endocytosis, macropinocytosis and phagocytosis47,51–55. However, it remains to be determined as to which EV uptake mechanism(s) is employed in different cell types under various conditions. The route of EV uptake is likely dependent on the following factors: (a) lipid and protein composition of the EV and plasma membranes of donor and recipient cell types; (b) EV subtypes; (c) state of cells releasing and taking up EVs, e.g. healthy or diseased; and (d) conditions in the extracellular space, e.g. acidic vs. basic, hypoxic, extracellular matrix components, etc.. The study presented here provides a means of successfully monitored EV docking and uptake, which will allow elucidation of these release and uptake mechanisms. It should also be noted that inhibition of protein translation in recipient cells by CHX treatment did not appear to affect EV uptake in this experimental setup. We speculate this could be a result of 1) uniform attachment of EVs by centrifugation at 4 °C prior to the CHX treatment; 2) pre-existing proteins available for the subsequently internalization of EVs; and 3) docked EV-cell membrane fusion that can readily occur when the temperature resumed to 37 °C following the centrifugation. Future studies will provide insight as to whether nascent protein synthesis is required for EV uptake via endocytosis and/or membrane fusion.
We further demonstrated that, under our experimental setup, EV-mediated RNA delivery can induce NFkB activity over an extended period of time (up to 72 h) in the recipient cells in a dose-dependent manner. EVs from normal and tumor cells are capable of modulating the immune response, as well as the cellular physiology in the tumor microenvironment56–59. And here we show direct evidence that EV-RNA can impact major signaling pathways in recipient cells such as NFkB, a key mediator of the immune response as well as inflammation and tumorigenesis60. While the physiological relevant EV dose and treatment duration remains to be defined, the current findings suggest that uptake of EVs by recipient cells in a microenvironment and at distal sites under pathological conditions, such as cancer, could result in immediate and systemic NFkB responses, respectively.
Our findings support EV-mediated communication between cells, which can be complex, multi-directional, and far-reaching. Protein and RNA cargos can not only be transferred to elicit phenotypic changes in recipient cells, but foreign mRNAs contained within EVs can also be rapidly translated. This latter finding supports potential RNA therapeutic applications of vesicles5. Future studies should focus on the dynamics of EV biogenesis, release and uptake considering EVs as a multifaceted communication platform between local and distant cell populations.
METHODS
Cell Culture
Human embryonic kidney 293T cells [American Type Culture Collection (ATCC), Manassas, VA], Gli36 glioma cells (Dr. Anthony Capanogni, UCLA, Los Angeles, CA), and primary GBM cells 20/37 were cultured in high glucose Dulbecco’s modified Eagle’s medium (Corning Cellgro, Manassas, VA) containing 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO) and 100 U per ml penicillin, 100 μg per ml streptomycin (Invitrogen, Grand Island, NY) in a humidified atmosphere at 37°C, 5% CO2. EL4 cells (ATCC) were maintained in RPMI 1640 adjusted to contain 1.5 g per L sodium bicarbonate, 4.5 g per L glucose, 10 mM HEPES, 1.0 mM sodium pyruvate, and supplemented with 0.05 mM 2-mercaptoethanol and 10% fetal bovine serum.
Reporter Constructs
Palmitoylation sequences (MLCCMRRTKQ) of growth cone-associated protein (GAP43)61 was genetically fused to the NH2-terminus of GFP (PalmGFP) and tdTomato (PalmtdTomato) by PCR using Phusion® High-Fidelity DNA Polymerase (New England BioLabs Inc., Ipswich, MA, USA). Plasmids pCAG-mGFP (a gift from Connie Cepko; Addgene plasmid 11150)62 and pCSCMV:tdTomato (a gift from Gerhart Ryffel; Addgene plasmid 30530)63 were used as cDNA templates for GAP-43, EGFP, and tdTomato (Supplementary Table 1 for primers). PalmGFP and PalmtdTomato sequences were inserted at NheI and XhoI sites of CSCGW2 lentivector plasmid kindly provided by Dr. Miguel Sena-Esteves (University of Massachusetts Medical School, Worcester, MA, USA)64, tagged with MS24X repeats between PspOMI and HpaI sites followed by β-actin 3′ UTR (a kind gift from Dr. Robert Singer, Albert Einstein College of Medicine, Bronx, NY, USA) between HpaI and XhoI sites. EV-GlucB construct was used to label EVs with a membrane-bound version of Gluc as previously described13.
EV Production and Isolation
Cells were transfected with DNA constructs using polyethyleneimine65, siGLO RISC-free siRNA with Lipofectamine 2000 (Invitrogen), or stably transduced with packaged lentivirus vectors (MGH Vector Core, Boston, MA, USA) to express genes of interest. EVs were isolated, as previously described13. Briefly, conditioned medium was collected from cells incubated with culture medium supplemented with 10% EV-depleted FBS for 48 h, centrifuged at 300 g for 10 min, 2,000 g for 10 min at 4°C, and filtered through a 0.8 μm (or 0.22 μm as specified) filter (Millipore, Billerica, MA, USA) before ultracentrifugation at 100,000 g for 90 min at 4°C. EV pellets were resuspended with double-0.22 μm-membrane filtered PBS.
Live-Cell Confocal Microscopy
Micrographs were captured with an LSM510 confocal microscope equipped with either 63X Zeiss Plan-APOCHROMAT oil, 1.4NA or 100X Zeiss Plan-APOCHROMAT DIC oil, 1.46NA under environmental control at 37°C and 5% CO2 (Zeiss, Thornwood, NY, USA).
EV Semi-quantification
Isolated EVs were first mounted onto a microscope slide with a #1 circular coverglass, allowed to settle for 30 min at room temperature, and imaged by confocal microscopy. To semi-quantitate EVs, EV signals in the confocal micrographs were first digitally displayed as spheres and then semi-quantitated by Imaris software (Bitplane, South Windsor, CT, USA). Same threshold for EV digital display and semi-quantification was applied to all samples. For semi-quantification of Palmtdtomato-EV and PKH67 signals, confocal micrographs were analyzed by MCID software (MCID Image Analysis Software Solutions for Life Sciences, USA).
Sucrose Density Gradients and Western Blot Analysis
Isolated EVs were subjected to sucrose density gradients consisting of 8, 30, 45 and 60% layers in PBS and centrifuged at 232,500 g for 30 min at 4°C. Following removal of the top layer, 10 fractions were collected and diluted 1:10 in PBS followed by centrifugation at 100,000 g to collect EV-containing pellets.
The pellets were lysed with 25 μl RIPA buffer supplemented with protease inhibitor (mini, Complete, Roche Diagnostics, Indianapolis, IN, USA), resolved by 10% SDS-PAGE with molecular weight standards (Precision Plus Protein All Blue Standards, Bio-Rad, Hercules, CA, USA), and transferred onto nitrocellulose membranes. The membranes were blocked with 5% bovine serum albumin (BSA) fraction V (Invitrogen), immunoblotted with anti-GFP (mouse; Invitrogen), anti-dsRed (to detect tdTomato; rabbit; Clontech, Mountain View, CA, USA) and anti-Alix (mouse; Santa Cruz, Dallas, TX, USA) antibodies followed by secondary antibodies conjugated to horseradish peroxidase (HRP; Molecular Probes, Eugene, OR, USA). Signal was detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA).
Transmission Electron Microscopy and Immunogold Staining
Isolated EVs were pelleted at 20,000 g for 30 min at 4°C, fixed with 4% formaldehyde in PBS for 2 h, washed in PBS and embedded in 20% gelatin. The gelatin embedded pellet was infiltrated in 2.3M sucrose (cryoprotection) and frozen in liquid nitrogen (LN2). Ultrathin (approximately 80 nm) frozen sections were cut at −120°C and picked up on formvar/carbon coated copper grids. The thawed sections were immunolabeled with anti-GFP (rabbit, AbCam #6556) or anti-dsRed (mouse; Clontech) followed by 5 or 10 nm protein A-gold secondary antibodies (University Medical Center, Utrecht, The Netherlands), respectively. Images were captured using Tecnai G2 Spirit Bio TWIN transmission electron microscope.
Multiphoton Intravital Microscopy (MP-IVM) of mouse tumors
Dorsal skinfold chambers (DSFCs) were installed on C57BL6 (B6) female mice (The Jackson Laboratory, Bar Harbor, ME), as previously described37,38. One day later, 106 EL4, EL4-PalmGFP, or EL4-GFP tumor cells in 50 μl HBSS were implanted into the chambers by injection into the skin tissue. Nine days later, animals were anaesthetized by intraperitoneal injection of Ketamine and Xylazine, and positioned on a custom-made microscopy stage that permitted maintaining the temperature of exposed skin tissue at 37°C. Purified EVs from EL4-PalmGFP cells were injected into tumors through the intact skin from the side opposing the exposed skin surface. All experiments were in accordance with NIH guidelines and were approved by the Institutional Animal Committees of Massachusetts General Hospital.
EV Uptake Analysis and EV-RNA Translation Assay
For the transwell setup to assess the diffraction limit and sensitivity of the microscope on EVs, cell culture inserts with 1 μm pore (BD Biosciences, San Jose, CA, USA) in a 24-well plate placed with coverslips within the wells were used.
For EV donors, 293T cells were infected with lentiviruses to stably both express PalmGFP and GlucB reporters13. PalmGFP+GlucB+ EVs were isolated from 0.8 μm filtered conditioned medium of the transduced cells, as described above. For EV recipient cells, Gli36 cells were stable transfected to express mCherry reporter (Gli36-mCherry).
To allow uniform docking of EVs and halt EV uptake by the recipient cells, Gli36-mCherry cells in a 24-well plate were treated with PalmGFP+GlucB+ EVs in EV-depleted medium and centrifuged at 1,800 rpm (maximum rotor radius: 19.2 cm) at 4°C for 90 min. PalmGFP+GlucB+ EV-containing medium was then removed, and the cells were washed once with PBS followed by replacement with EV-depleted medium with or without cycloheximide (CHX; 20 μg per ml). At the time of centrifugation (−1.5 h), 0, 1, 3, 6, 12 and 24 h post-centrifugation, conditioned medium was first collected, and cells were washed with PBS followed by trypsinization with 0.05% trypsin/0.53 mM EDTA (Corning Cellgro) and resuspension in PBS for flow cytometry analysis on a BD LSRII Multi-Laser Analyzer (BD Biosciences) to determine EV uptake by the recipient cells.
To assess translation of EV-delivered GlucB RNA, the collected cells and conditioned medium were plated in triplicates into a white 96-well luminometer plate. GlucB activity was then measured by an MLX Microtiter plate luminometer (Dynex Technologies, Chantilly, VA, USA) with automated injection of 50 μl coelentrazine (CTZ; 8 ng per mL; Nanolight, Pinetop, AZ).
Nascent Protein Synthesis Assay
To detect CHX-mediated inhibition of protein synthesis, Gli36-mCherry cells in a 24-well plate were prepared in parallel to the EV-uptake and –RNA translation experiment as described above. At the indicated post-centrifugation time points, culture medium was replaced with methionine- and cysteine-free medium containing Click-iT® homopropargylglycine (HPG; Invitrogen) for 30 min, and the cells were washed with PBS, trypsinized with 0.05% trypsin/0.53 mM EDTA (Corning Cellgro), resuspended in PBS, pelleted at 1,500 rpm (maximum rotor radius: 20.78 cm) for 5 min at 4°C, and resuspended in 80% ethanol for 15 min on ice. The fixed cells were conjugated with Alexa Fluor® 488 azide to reveal HPG-labeling, and stained with NuclearMask™ Blue stain according to manufacture’s protocol (Invitrogen). The labeled cells were resuspended in PBS for flow cytometry analysis on a BD LSRII Multi-Laser Analyzer (BD Biosciences).
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA069246 (X.O.B., B.A.T. and R.W.), U19CA179563 (supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director; X.O.B. and T.R.M.), Voices Against Brain Cancer (X.O.B. and C.P.L.), and CA150975, CA179563, and AI073457 (T.R.M.). C.P.L was supported by the Canadian Institutes of Health Research (CIHR). We would like to thank D. Morse and K. Conway for the production of lentivirus at the MGH Vector Core, Charlestown, MA, USA supported by NIH/NINDS P30NS045776 (X.O.B. and B.A.T.); I. Armata, K.E. van der Vos and O. Mardini for technical assistance; M. Ericsson for electron microscopy (Conventional Electron Microscopy Core, Harvard Medical School, Boston, MA); T. Diefenbach for microscope assistance (Ragon Institute Microscopy Core, Cambridge, MA, USA); and R. Mylvaganam and C. Luo for flow cytometry assistance (MGH FACS Core, Charlestown, MA, USA supported by NIH 1S10RR023440-01A1).
Footnotes
AUTHOR CONTRIBUTIONS
C.P.L. generated and analyzed data for the fluorescent EV and EV-RNA reporters, confocal microscopy, sucrose gradient fractionation, dot blot assay, EV uptake and EV-RNA translation assays, and was involved in the experimental design and strategy and writing/editing of the manuscript. E.Y.K and T.R.M. designed, generated and analyzed MP-IVM data, and were involved in the writing/editing of the manuscript. C.E.B. and C.P.L. generated and analyzed NFkB data, and edited the manuscript. R. W., B.A.T. and X.O.B. were involved in experimental strategy, design, data analysis, and writing/editing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Colombo M, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. Journal of Cell Science. 2013 doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 2.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney International. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 3.Vader P, Breakefield XO, Wood MJA. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol Med. 2014 doi: 10.1016/j.molmed.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 5.Lai CP, Breakefield XO. Role of exosomes/microvesicles in the nervous system and use in emerging therapies. Frontiers in Physiology. 2012;3:228. doi: 10.3389/fphys.2012.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 9.Koumangoye RB, Sakwe AM, Goodwin JS, Patel T, Ochieng J. Detachment of Breast Tumor Cells Induces Rapid Secretion of Exosomes Which Subsequently Mediate Cellular Adhesion and Spreading. PLoS ONE. 2011;6:e24234. doi: 10.1371/journal.pone.0024234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B, Fang Y, Wu N, Gould SJ. Biogenesis of the Posterior Pole Is Mediated by the Exosome/Microvesicle Protein-sorting Pathway. Journal of Biological Chemistry. 2011;286:44162–44176. doi: 10.1074/jbc.M111.274803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suetsugu A, et al. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude mouse models. Advanced Drug Delivery Reviews. 2012:1–8. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, et al. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4:1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai CP, et al. Dynamic Biodistribution of Extracellular Vesicles In VivoUsing a Multimodal Imaging Reporter. ACS Nano. 2014;140102104824009 doi: 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekström K, et al. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. Journal of Extracellular Vesicles. 2012;1 doi: 10.3402/jev.v1i0.18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triola G, Waldmann H, Hedberg C. Chemical biology of lipidated proteins. ACS Chem Biol. 2012;7:87–99. doi: 10.1021/cb200460u. [DOI] [PubMed] [Google Scholar]
- 16.Zacharias DA. Partitioning of Lipid-Modified Monomeric GFPs into Membrane Microdomains of Live Cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 17.Zuber MX, Strittmatter SM, Fishman MC. A membrane-targeting signal in the amino terminus of the neuronal protein GAP-43. Nature. 1989;341:345–348. doi: 10.1038/341345a0. [DOI] [PubMed] [Google Scholar]
- 18.Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T, Gu F, Gruenberg J. Lipids, lipid domains and lipid-protein interactions in endocytic membrane traffic. Seminars in Cell & Developmental Biology. 1998;9:517–526. doi: 10.1006/scdb.1998.0257. [DOI] [PubMed] [Google Scholar]
- 20.Dolo V, et al. Enrichment and localization of ganglioside G(D3) and caveolin-1 in shed tumor cell membrane vesicles. Biochim Biophys Acta. 2000;1486:265–274. doi: 10.1016/s1388-1981(00)00063-9. [DOI] [PubMed] [Google Scholar]
- 21.Cocucci E, Racchetti G, Podini P, Meldolesi J. Enlargeosome Traffic: Exocytosis Triggered by Various Signals Is Followed by Endocytosis, Membrane Shedding or Both. Traffic. 2007;8:742–757. doi: 10.1111/j.1600-0854.2007.00566.x. [DOI] [PubMed] [Google Scholar]
- 22.Badr CE, Tannous BA. Bioluminescence imaging: progress and applications. Trends in Biotechnology. 2011;29:624–633. doi: 10.1016/j.tibtech.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannous BA, Kim D-E, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 25.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 26.Lee YS, Kim SH, Cho JA, Kim CW. Introduction of the CIITA gene into tumor cells produces exosomes with enhanced anti-tumor effects. Exp Mol Med. 2011;43:281–290. doi: 10.3858/emm.2011.43.5.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thery C, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 28.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang X, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Molecular Therapy. 2011;19:1769–1779. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 31.Silva AKA, et al. Magnetic and photoresponsive theranosomes: translating cell-released vesicles into smart nanovectors for cancer therapy. ACS Nano. 2013;7:4954–4966. doi: 10.1021/nn400269x. [DOI] [PubMed] [Google Scholar]
- 32.Ohno S-I, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Molecular Therapy. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace PK, et al. Tracking antigen-driven responses by flow cytometry: Monitoring proliferation by dye dilution. Cytometry. 2008;73A:1019–1034. doi: 10.1002/cyto.a.20619. [DOI] [PubMed] [Google Scholar]
- 34.Davis DM, Sowinski S. Membrane nanotubes: dynamic long-distance connections between animal cells. Nat Rev Mol Cell Biol. 2008;9:431–436. doi: 10.1038/nrm2399. [DOI] [PubMed] [Google Scholar]
- 35.Di Vizio D, Kim J, Hager M, Morello M, Yang W. Oncosome Formation in Prostate Cancer: Association with a Region of Frequent Chromosomal Deletion in Metastatic Disease. Cancer Research. 2009 doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atai NA, et al. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J Neurooncol. 2013;115:343–351. doi: 10.1007/s11060-013-1235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alieva M, Ritsma L, Giedt RJ, Weissleder R, van Rheenen J. Imaging windows for long-term intravital imaging. IntraVital. 2014;3:e29917. doi: 10.4161/intv.29917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marangoni F, Murooka TT, Manzo T, Kim EY. The Transcription Factor NFAT Exhibits Signal Memory during Serial T Cell Interactions with Antigen-Presenting Cells. Immunity. 2013 doi: 10.1016/j.immuni.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer CA, et al. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J Clin Invest. 2014;124:2425–2440. doi: 10.1172/JCI66375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badr CE, et al. Suicidal gene therapy in an NF-κB-controlled tumor environment as monitored by a secreted blood reporter. Gene Ther. 2011;18:445–451. doi: 10.1038/gt.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badr CE, et al. Real-time monitoring of nuclear factor kappaB activity in cultured cells and in animal models. Mol Imaging. 2009;8:278–290. [PMC free article] [PubMed] [Google Scholar]
- 43.Mittelbrunn MIA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Communications. 2011;2:282–10. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Das K, Woo J, Gimzewski JK. Nanofilaments on glioblastoma exosomes revealed by peak force microscopy. J R Soc Interface. 2014;11:20131150. doi: 10.1098/rsif.2013.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rieck B. Unexpected durability of PKH 26 staining on rat adipocytes. Cell Biology International. 2003;27:445–447. doi: 10.1016/s1065-6995(03)00036-2. [DOI] [PubMed] [Google Scholar]
- 46.Im H, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nature Biotechnology. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Meth. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parolini I, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson KJ, et al. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288:17713–17724. doi: 10.1074/jbc.M112.445403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morelli AE, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 53.Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. Exosomes Derived from Epstein-Barr Virus-Infected Cells Are Internalized via Caveola-Dependent Endocytosis and Promote Phenotypic Modulation in Target Cells. Journal of Virology. 2013;87:10334–10347. doi: 10.1128/JVI.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fitzner D, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of Cell Science. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 55.Feng D, et al. Cellular Internalization of Exosomes Occurs Through Phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 56.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 57.Valenti R, et al. Tumor-Released Microvesicles as Vehicles of Immunosuppression. Cancer Research. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 58.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Research. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 59.Luga V, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 60.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCabe JB, Berthiaume LG. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol Biol Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waldner C, Roose M, Ryffel GU. Red fluorescent Xenopus laevis: a new tool for grafting analysis. BMC Dev Biol. 2009;9:37. doi: 10.1186/1471-213X-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. Journal of Virological Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 65.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.