Figure 1.
A transgenic library of conditional miRNA competitive inhibitors.
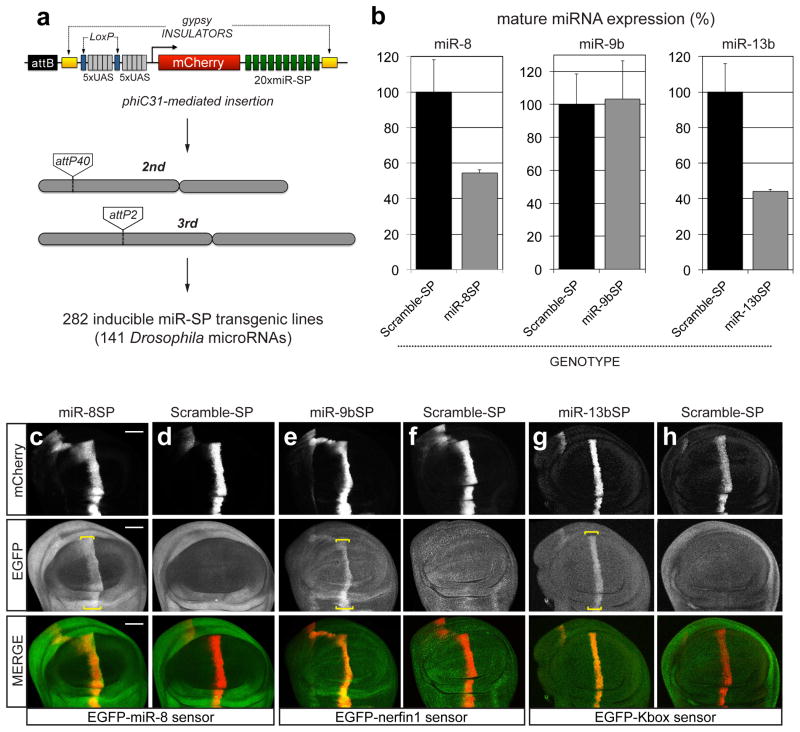
(a) Second-generation SP elements consist of twenty miRNA binding sites with mismatches at positions 9–12 placed in the 3′UTR of mCherry under the control of 10 tunable Gal4 UAS binding sites. The entire cassette was cloned in an attB vector containing gypsy insulators. phiC31-mediated positional integration was used to generate a library of 282 inducible lines covering 141 high confidence Drosophila miRNAs, at defined landing sites on the 2nd (attP40) and 3rd (attP2) autosomes. (b) Quantification of endogenous miR-8, miR-9b and miR-13b mature miRNA levels using Taqman qPCR in third instar larvae following ubiquitous expression (tubulin-Gal4) of corresponding miR-SP constructs compared to Scramble controls (Student’s t-test: 13b P=0.02; error bars, sem, n =3 biological replicates with 10 animals per sample). (c–h) Targeted expression of miR-8SP (c, Scale bar is 50μm), miR-9bSP (e) and miR-13bSP (g) with ptc-Gal4 in wing imaginal discs ubiquitously expressing tubulinEGFP-miR-8, tubulinEGFP-nerfin1 and tubulinEGFP-Kbox sensors respectively. Tissue-specific up-regulation of sensor levels was observed in cells along the anterior-posterior boundary of the disc. No change was apparent following expression of a Scramble-SP control (d, f, h).