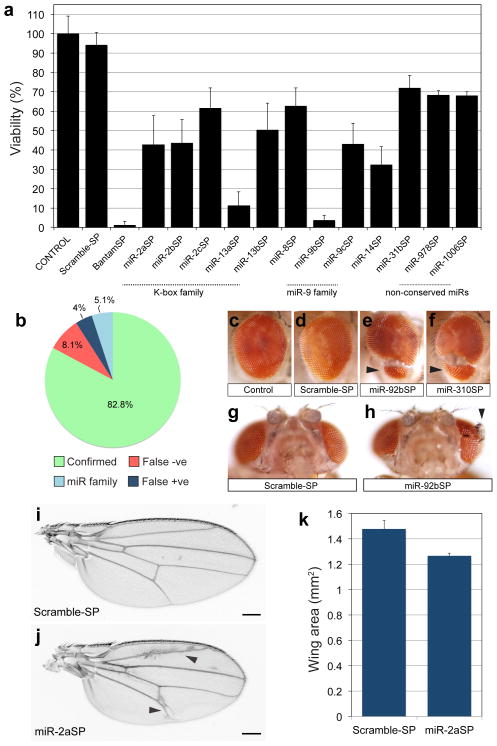
Figure 2.
Drosophila miRNA phenotypes in viability and external morphology. (a) Viability defects following ubiquitous expression of SP library under the tubulin-Gal4 driver. Data is displayed as average percent viability relative to control; at least six independent replicate batches were analyzed for each genotype (ANOVA, post hoc analysis with Tukey-Kramer multiple comparisons test P≤ 0.01; error bars, s.d). (b) Benchmark comparison of viability phenotypes to Drosophila miRNA null mutants13. “Confirmed” indicates same viability phenotype shared with SP and null. False negative (“False −ve”) indicates miRNAs where null demonstrated viability impaired phenotypes, but miR-SP lines were viable. False positive (“False +ve”) indicates miRNAs where a phenotype was observed with miR-SP but not in the null animal. “miR family” represents miRNAs for which a similar seed sequence is shared and miRSP lines display a viability phenotype that is only confirmed by an individual family member. The denominator for this chart was 99; we excluded all lines for which there was no null available, lines that were not tested by Chen and colleagues13, mutants removing entire clusters of multiple miRs, or mutants for which complementation was inconclusive (Supplementary Data 3). (c-h) Eye morphology defects (arrowheads) following inhibition of miR-92 activity. Genotypes: +/tubulin-Gal4 (c), +/Scramble-SP;tubulin-Gal4/Scramble-SP (d, g), +/miR-92bSP;tubulin-Gal4/miR-92bSP (e, h) +/miR-92bSP;tubulin-Gal4/miR-310SP (f). (i–k) Expression of miR-2aSP results in wing vein patterning abnormalities (j, arrowheads) and an overall reduction in wing blade size (j, Scale bar is 200μm). Average wing size area was quantified in triplicate samples (P=0.004; error bars, sem) from +/Scramble-SP;tubulin-Gal4/Scramble-SP and +/miR-2aSP;tubulin-Gal4/miR-2aSP animals (k).