Abstract
RNA interference (RNAi) is a powerful tool for the regulation of gene expression. Small exogenous noncoding RNAs (ncRNAs) such as siRNA and shRNA are the active silencing agents, intended to target and cleave complementary mRNAs in a specific way. They are widely and successfully employed in functional studies, and several ongoing and already completed siRNA-based clinical trials suggest encouraging results in the regulation of overexpressed genes in disease.
siRNAs share many aspects of their biogenesis and function with miRNAs, small ncRNA molecules transcribed from endogenous genes which are able to repress the expression of target mRNAs by either inhibiting their translation or promoting their degradation. Although siRNA and artificial miRNA molecules can significantly reduce the expression of overexpressed target genes, cancer and other diseases can also be triggered or sustained by upregulated miRNAs.
Thus, in the past recent years, molecular tools for miRNA silencing, such as antagomiRs and miRNA sponges, have been developed. These molecules have shown their efficacy in the derepression of genes downregulated by overexpressed miRNAs. In particular, while a single antagomiR is able to inhibit a single complementary miRNA, an artificial sponge construct usually contains one or more binding sites for one or more miRNAs and functions by competing with the natural targets of these miRNAs. As a consequence, natural miRNA targets are reexpressed at their physiological level.
In this chapter we review the most successful methods for the computational design of siRNAs, antagomiRs, and miRNA sponges and describe the most popular tools that implement them.
Keywords: RNAi, siRNA, shRNA, antagomiR, miRNA, Sponge, Gene expression
1 Introduction
The discovery of the first short ncRNA capable of acting as endogenous regulator of gene expression was made by Ambros et al. in 1993, while investigating the role of lin-4 in the developmental timing of C. elegans [1]. That accomplishment revealed only a glimpse of the much broader reality uncovered by Fire and Mello 5 years later when they reported the capability of exogenous double-stranded RNAs to silence genes in a specific manner, disclosing the mechanism we know today as RNA interference (RNAi) [2]. In 1999, a similar process was discovered to take place in plants, as short RNA sequences (~20–25 nt) were found to be able to bind their mRNA targets through perfect base complementarity [3]. Since then, a revolution has occurred in the field of RNA biology, thanks to the characterization of RNAi as an innate biological process through which the expression of specifically targeted genes can be modulated and/or silenced, ushering in a new world of research and potential therapeutic applications. Specifically, RNAi is a naturally occurring mechanism resulting in a sequence-specific, posttranscriptional downregulation of gene expression induced by double-stranded RNA (dsRNA) homologous to the target gene. This regulation can occur at different levels of the gene expression process, including transcription, mRNA processing, and translation. The small RNA molecules responsible for RNAi originate from endogenous and exogenous double-stranded precursors and play a specific role in guiding effector protein complexes toward their nucleic acid targets by partial or full complementarity bonds [4].
Although several classes of regulatory ncRNAs have been identified, the most relevant ones can essentially be represented by two groups, according to their origin, structure, associated effector proteins, and function: small interfering RNAs (siRNAs), principally of exogenous origin in animals, and microRNAs (miRNAs), which are endogenous genome products. These molecules, apparently present only in eukaryotes and in some viruses, are in fact the most abundant regulatory molecules in terms of both phylogeny and physiology.
siRNA is a class of double-stranded RNA molecules, 20–25 base pairs in length, whose role appears to be involved in the defense of the cell and maintenance of genome integrity through the silencing of exogenous nucleic acids and undesired transcripts (such as transposons and repetitive elements) [4, 5]. Although their posttranscriptional gene-silencing role in plants and in some simple animal species was shown to be of endogenous origin, siRNAs in animals, instead, are mainly exogenous molecules obtained from perfectly complementary long double-stranded precursors coming from viruses and transposons [3]. As it was first demonstrated in nematodes [2] and later in mammalians [6], siRNA-mediated RNAi can be obtained either by simple dsRNAs of about 21 nucleotides (nt) with two-nucleotide 3′ overhang or by stably expressed short hairpin RNAs (shRNAs), which are processed into siRNAs [7, 8]. Compared to unprocessed siRNA, this last approach undoubtedly presents many advantages, such as long-lasting silencing effects, cost-effectiveness, as well as easy delivery methods. Generally, shRNA is transcribed in cells from a DNA template as a single-stranded RNA molecule (about 50–100 bases). The complementary regions are spaced by a hairpin loop, therefore the name “short hairpin” RNA [9]. Intracellular presence of siRNAs which are perfectly complementary to their target mRNAs is of crucial importance for the induction of RNAi and leads to mRNA degradation.
While siRNAs are mainly exogenous molecules, miRNAs instead are a class of RNAi inducers which derive from partially complementary double-stranded hairpin precursors of endogenous origin. Once processed, they are small single-stranded RNAs (20–22 nt long) able to modulate posttranscriptional gene silencing through repression, and at times degradation, of specific mRNA target molecules [10]. It has been estimated that miRNA-coding genes represent 1 % of the total gene population, being the biggest class of regulatory molecules. They are present in plants, higher eukaryotes, and in some viruses. miRNAs are often encoded in clusters by genes usually located in introns and, more rarely, in exons of protein-coding genes, as well as in intergenic regions [10]. RNA polymerases produce primary miRNA transcripts (primiRNAs) from microRNA genes [11–13]. pri-miRNAs are approximately >100 nt long and are subsequently processed into ~70-nt precursor miRNAs (pre-miRNAs) by a microprocessor complex comprised of the enzyme Drosha and a subunit DCGR8 [14–16]. Pre-miRNAs are then exported into the cytoplasm by the exportin 5 protein [17, 18]. From here on, in spite of their originating difference, common features, such as the length of their mature products and their sequence-specific inhibitory functions, suggest that siRNA and miRNA have similar processing and common mechanisms. Their double-stranded precursors (shRNAs and pre-miRNAs) are indeed both cleaved by Dicer, a ribonuclease III-type protein [9, 19–21], into short 19–21 duplexes having two symmetric nucleotide overhangs at the 3′ end and a 5′ phosphate along with a 3′ hydroxyl group. After cleavage, Dicer, together with protein complexes TRBP and PACT, loads the obtained duplexes into a nuclease-containing multiprotein complex referred to as the RNA-induced silencing complex (RISC). Once the duplex is loaded, the Ago2 protein component of the RISC then cleaves one of the two strands of the duplex, which is thus, by convention, considered the sense “passenger” strand. The antisense strand, which remains loaded into the thus activated RISC, is instead called “guide” since it acts as an adapter for the complex to mRNA targets and allows it to carry out the RNAi mechanism [22–26]. In miRNAs, the strand which most commonly plays the role of the guide is called “mature miRNA,” while the other one is called “miRNA*” [27]. Animal 3′ UTR sequences often present miRNA binding sites in multiple copies, while, conversely, most miRNAs in plants, as well as siRNAs, bind their targets in their coding regions with perfect complementarity. Another fundamental difference between miRNAs and siRNAs consists, in fact, in the type of binding, which is considered a key factor in their regulatory function: miRNAs bind their target with partial complementarity, allowing bulges and loops in duplexes. However, a key feature in their target recognition is represented by the perfect base pairing with the target in positions 2–8 of the miRNA guide, which is known as the seed region. The presence of mismatches in the central part of the duplex is usually associated to translational repression, which seems to be the default mechanism of miRNA-mediated RNAi. The cleavage of perfectly paired duplexes, which is the default RNAi mechanism in the case of siRNAs, is instead considered for miRNAs an additional feature leading to the same effect on the protein level.
The biological role of these molecules is currently being intensively elucidated, and their involvement in fundamental processes, such as apoptosis, metabolism, cell proliferation, and organism development, has been widely demonstrated.
Due to the fast and cost-effective way of disrupting genes’ functions provided by RNAi-based gene knockdown techniques, great and rapid progress has been made in recent years, and siRNAs have become a standard tool routinely used in molecular genetics and functional genomics laboratory [28]. Moreover, a large number of RNAi-based potential therapeutic agents are actively being explored, while several RNAi-based therapies against several diseases such as viral infections, inflammatory diseases, and cancers have already reached preclinical and even clinical stage in development [29–31].
In light of all this, great interest has thus eventually arisen in loss-of-function studies in vivo in order to further investigate the precise molecular function of miRNAs in mammals, which is still unknown on the greater part. Thus in 2005, Krutzfeldt et al. devised a novel class of chemically engineered oligonucleotides able to silence endogenous miRNAs in vivo, which they termed “ antagomiRs” [32]. These new molecules were also shown to perform efficient and stable loss-of-function phenotypes for specific miRNAs by lentivirus-mediated delivery in cultured cells [33]. In 2007, other miRNA inhibitors, called “miRNA sponges,” were devised [34]. They consisted in transcripts expressed from strong promoters and essentially containing multiple, tandem binding sites to an miRNA of interest. Once vectors encoding these competitive miRNA inhibitors were transiently transfected into cultured cells, miRNA targets were shown to be derepressed just as strongly as previously accomplished, thus suggesting a valid alternative to antagomiRs.
These results clearly show that these molecular constructs may represent a valid strategy for silencing miRNA in diseases, such as cancer, and further investigate potential therapeutic applications.
In this chapter we will give an overview of the most successful approaches and algorithms regarding RNAi design techniques and briefly describe the most popular tools that implement them.
2 Materials
The methods described in this chapter are implemented in tools publicly available online. They can be executed on any personal computer equipped with Internet connection and a browser and don’t require any particular resource.
3 Methods
RNAi represents today a well-established approach for gene silencing in loss-of-function studies and genetic screens. Since its discovery in 1998, the main focuses of RNAi computational research have been the discovery of siRNA design rules and the development of siRNA efficacy and specificity prediction models. Nevertheless, a considerable number of siRNA design tools and siRNA databases are freely available online and widely employed for gene knockdown experiments.
The other side of artificial RNAi is represented by molecular tools for miRNA silencing, such as antagomiRs and sponges.
In this section we summarize the main rules for the design of siRNA, antagomiRs, and sponges and provide a brief introduction to several tools and databases which are freely available online.
3.1 Design of Functional siRNA
From a computational point of view, siRNA design is the process of choosing a functional binding site on a target mRNA sequence, which will correspond to the sense strand of the siRNA under design (typically 21–23 nt long) [35]. The siRNA antisense sequence is obtained as the complement to the sense strand. Symmetric 3′ overhangs, usually dTdT, are added to improve stability of the duplex and to facilitate RISC loading, ensuring equal ratios of sense and antisense strands incorporation [6, 36, 37]. Other overhang sequences are acceptable, but some combinations, such as GG, should be avoided. The efficacy of RNAi is mostly determined by sequence-specific factors which affect the stability of the duplex ends. siRNA duplexes often have asymmetric loading of the antisense versus sense strands [38, 39]. The strand whose 5′ end is thermodynamically less stable is preferentially incorporated into the RISC.
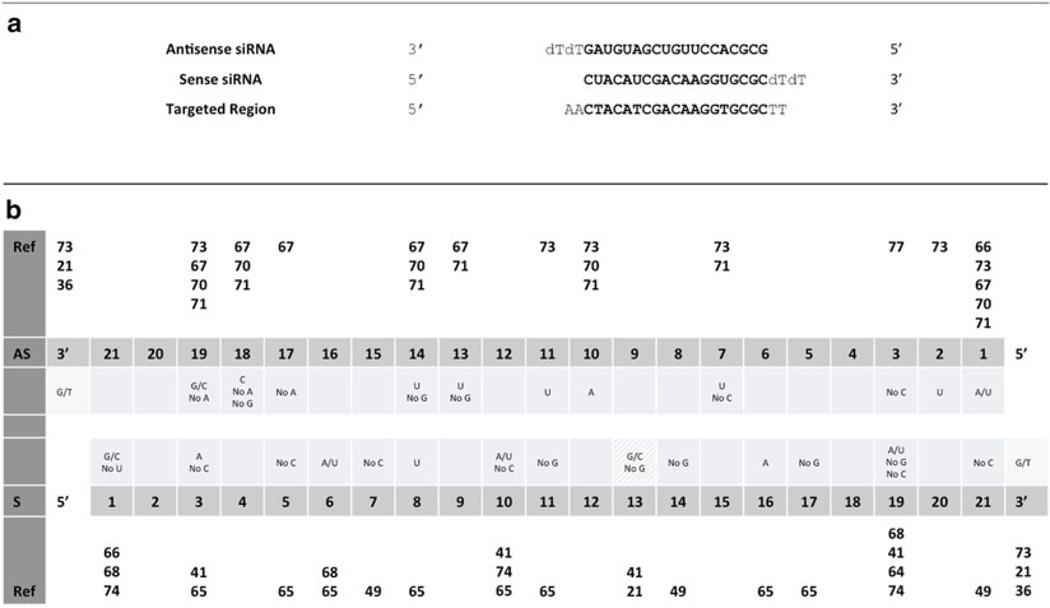
Elbashir et al. suggest to choose the 23-nt sequence motif AA(N19)TT as binding site (N19 means any combination of 19 nucleotides), where (N19)TT corresponds to the sense strand of the siRNA, while the complement to AA(N19) corresponds to the antisense strand (see Fig. 1a) [6].
Fig. 1.
siRNA design rules. (a) An example of target region in an mRNA sequence and the corresponding siRNA duplex with 3′ overhangs. (b) Specific positional rules for siRNA design. The darker cells represent positions on the siRNA antisense (AS) and sense (S) strands. The light gray cells contain specific rules for the corresponding positions. For each set of rules, references are given (Ref). The striped gray background indicates inconsistencies of the rules, due to the different experiments that they come from
Many different features associated to functional siRNAs have been identified in the past years and some of them are now widely accepted as standard rules in siRNA design and are implemented in the majority of design tools. They can be classified into four different categories: (1) general binding rules, (2) nucleotide composition rules, (3) specific positional rules, and (4) thermodynamics rules. Table 1 summarizes categories (1), (2), and (4), while rules in category (3) are represented in Fig. 1b.
Table 1.
Rules for siRNA design
| Design rule | References |
|---|---|
| General targeting rules | |
| Select the target region preferably 50–100-nt downstream of the start codon | [21] |
| Avoid to target the middle of the coding sequence of the target gene | [64] |
| Pooling of four or five siRNA duplexes per gene | [64] |
| Nucleotide composition rules | |
| Antisense strand with higher information content | [65] |
| Absence of any GC stretch >9 nt long | [66] |
| At least five A/U residues in the 5′ terminal one-third of the antisense strand | [49, 66, 67] |
| A higher “A/U” content in the 3′ end than that in the 5′ end (sense strand) | [68] |
| G/C content ranges: 32–58 %, 30–52 %, 32–79 %, 36–53 %, 35–73 %, 25–55 % | [21, 41, 68–71] |
| Absence of internal repeats | [41] |
| Presence of motifs “AAC,” “UC,” “UG,” “AAG,” “AGC,” “UCU,” “UCCG,” “CUU,” “CU,” “GUU,” “UCC,” “CG,” “AUC,” “GCG,” “UUU,” “ACA,” “UUC,” “CAA” in antisense strand | [49, 70–72] |
| Avoid motifs “CUU,” “CUA,” “GUU,” “GU,” “GAU,” “ACGA,” “GCC,” “GUGG,” “CCC,” “GGC,” “CCG,” “GGG,” “CAG,” “GAG,” “GCA,” “AUA,” “CUG,” “AG,” “GG,” “GGA” in antisense strand | [49, 70–72] |
| High content of “U” in antisense strand | [72] |
| Low content of “G” in antisense strand | [72] |
| Thermodynamics rules | |
| Total hairpin energy <1 | [69] |
| Antisense 5′ end binding energy <9 | [69] |
| Sense 5′ end binding energy in range 5–9 | [69] |
| Middle binding energy <13 | [69] |
| Energy difference <0 | [69] |
| Energy difference within −1 and 0 | [69] |
| Significant ΔG difference between positions 1 and 18 | [67] |
| High ΔG in positions 1–4, 5–8, and 13–14 in the antisense strand | [70] |
| Low ΔG in positions 18–19 in the antisense strand | [70] |
| Avoid folding of siRNA | [70] |
The rules are classified in four different categories, three of which are summarized in this table (see also Fig. 1b).
For each rule, or group of rules, references are given
General binding rules refer to factors such as the position of the binding sites in the target transcript. For example, the target region should preferably be between 50 and 100 nt downstream of the start codon, and the middle of the coding sequence should be avoided. Another rule suggests pooling of four or five siRNA duplexes per target gene, in order to ensure a stronger repression.
Another class of rules concerns the siRNA nucleotide composition. A major feature, implemented by every design tool, is the G/C content, which should typically be in the range of 30–55 %, although values as low as 25 % or as high as 79 % are still associated to functional siRNAs.
Other features in this category include the presence/absence of particular motifs in the antisense strand and the absence of internal repeats.
Specific positional rules are the most numerous and regard the selection of nucleotides to prefer or avoid in specific positions of either the sense or the antisense strand of the duplex. For example, the antisense sequence should always have an A/U base at its 5′ end. This is associated to a weaker thermodynamic stability which facilitates incorporation of the strand into the RISC, as already discussed above. Other rules suggest to avoid G/C nucleotides at the 5′ end of the sense strand or to have either a G or a C in position 4 of the sense strand.
Finally, thermodynamics rules refer to the global or local energy of the duplex and are partly related to the choice of nucleotides in specific positions such as the 5′ end of the antisense strand, as already mentioned, or in other regions such as the middle of the duplex. Other thermodynamic features associated to siRNA efficacy include the structural accessibility of the target site and the fact that folding of siRNAs should be avoided.
These and other rules are implemented in a considerable number of siRNA design tools available online, which will be described in the next subsection. One thing that is worth mention is that all the rules were obtained from distinct experiments performed in different conditions by different labs; thus inconsistencies and contradictions are inevitable, such as the one highlighted in Fig. 1b. This is one major drawback as discussed at the end of next section.
3.2 siRNA Design Online Resources
Table 2 provides a list of tools for the automated design of siRNA and shRNA sequences. Most tools have user-friendly interfaces which don’t require any additional specifications aside from the target sequence.
Table 2.
Tools for siRNA design
| Tool | Param | sh | as | Therm | Off | Link |
|---|---|---|---|---|---|---|
| OptiRNAi 2.0 | ✕ | ✕ | ✕ | ✕ | ✕ | http://rnai.nci.nih.gov |
| siDirect 2 | ✕ | ✕ | ✔ | ✔ | ✔ | http://sidirect2.rnai.jp |
| siRNA scales | ✕ | ✕ | ✔ | ✕ | ✕ | http://gesteland.genetics.utah.edu/siRNA_scales |
| siExplorer | ✕ | ✕ | ✔ | ✕ | ✔ | http://rna.chem.t.u-tokyo.ac.jp/cgi/siexplorer.htm |
| RFRCDB-siRNA | ✕ | ✕ | ✔ | ✕ | ✔ | http://www.bioinf.seu.edu.cn/siRNA/index.htm |
| OligoWalk | ✕ | ✕ | ✔ | ✔ | ✕ | http://rna.urmc.rochester.edu/cgi-bin/server_exe/oligowalk/oligowalk_form.cgi |
| Sfold | ✔ | ✕ | ✔ | ✔ | ✕ | http://sfold.wadsworth.org |
| siMAX | ✔ | ✕ | ✔ | ✔ | ✔ | http://www.operon.com/products/siRNA/sirna-overview.aspx |
| DSIR | ✔ | ✔ | ✔ | ✕ | ✔ | http://biodev.cea.fr/DSIR/ |
| siRNA scan | ✔ | ✕ | ✔ | ✕ | ✔ | http://bioinfo2.noble.org/RNAiScan.htm |
| RNAxs | ✔ | ✕ | ✔ | ✔ | ✔ | http://rna.tbi.univie.ac.at/cgi-bin/RNAxs |
| i-Score | ✔ | ✕ | ✔ | ✔ | ✕ | http://www.med.nagoya-u.ac.jp/neurogenetics/i_Score/i_score.html |
| siVirus | ✔ | ✕ | ✕ | ✕ | ✔ | http://sivirus.rnai.jp |
Details and links are given for each tool. Param: it is possible to provide parameters and constraints as input. sh: the tool also returns the complete shRNA sequence. as: both antisense and sense siRNA sequences are returned. Therm: the tool takes into account thermodynamic features. Off: the tool performs or has the option for the computation of off-target sequences for the designed siRNAs
OptiRNAi 2.0 is a fast tool which predicts 21–23-nt RNAi target sites on a user-provided sequence using the criteria described by Elbashir et al. in 2001 and Reynolds et al. in 2004 [36, 40, 41]. The program generates a list of up to ten siRNA target sites for each of which a score indicates how well it matches the considered features. The tool doesn’t return the actual siRNA antisense sequence, which has to be manually derived as the reverse complement of the binding sites.
As for OptiRNAi, siDirect 2 also accepts just the target sequence as input. It returns a table with a list of potential binding sites (including the 2-nt overhang) [42]. For each site, the corresponding siRNA duplex strands are given, together with the melting temperature (Tm) of the seed-target duplex, as a measure of thermodynamic stability. The seed-target duplex is formed between the region 2–8 of the siRNA guide strand (from the 5′ end) and its target mRNA site.
Other details provided include the list of potential off-target genes for the guide and passenger strands and a graphical view of the siRNA binding sites in the target sequence.
siRNA scales are another design tool which accepts as input a target sequence and returns a list with all possible 19-nt long siRNA sequences and the predicted percentage of target mRNA copies present in the cells after siRNA-directed cleavage as a measure of efficiency [43]. Both sense and antisense strands are returned. Users can also specify to show only siRNAs with high predicted efficiency (%mRNA ≤ 30).
siExplorer and RFRCDB-siRNA are other siRNA design tools which accept a target sequence as input and returns a list of potential binding sites and siRNAs ranked by their experimental or predicted efficiency [44, 45]. In particular, siExplorer also returns GC% content and, for each sequence, provides a link to perform a BLAST search for off-target evaluation. Moreover, charts with the distribution of prediction scores and the distribution of the top 10 binding sites on the target sequences are visualized. Users can choose to show the top 10, 20, or 50 results that can also be downloaded in Excel format. RFRCDB-siRNA also provides tested sequences in addition to the predicted ones. The tool, indeed, performs a database search on an experimentally validated siRNA database in order to find possible matches with the user-provided sequence.
OligoWalk is another siRNA design tool [46]. It returns a list of siRNA sequences with their probability of being efficient. For each siRNA, a thermodynamics analysis is performed. In particular, the target structure is computed before and after oligo binding, and details about the energy and the Tm of the duplex, together with other energy values, are provided.
Sfold is a suite of RNA folding prediction tools, which include a program for the design of siRNA sequences based on thermodynamics features [47]. The tool allows interactive computation for up to 250-nt-long target sequences and batch processing for longer sequences. In the latter case, a notification is sent by e-mail when results are available. In addition to the target sequence, users can also provide further structural constraint information, i.e., force/prevent pairing of specific base pairs.
Several files are returned as output, containing detailed information on the predicted siRNAs and their interaction with the target, such as the siRNA duplex GC content and thermodynamics score, the total stability of the duplex and the differential stability of its ends, the target accessibility score, the average internal stability and disruption energy of the binding site, and the sum of probabilities of unpaired target bases. Other details regarding secondary structure probabilities are also given and various filters are available. Finally, the complete probability profile, the regional probability profile, and the siRNA internal stability profile are provided as graph charts.
Other tools with more sophisticated interfaces allow the specification of parameters and constraints about the siRNA to be designed and its target sequence.
siMAX is a design resource whose input consists of the target sequence together with a limited number of parameters such as GC content range, minimum and maximum distance from start and stop codons, and the mRNA motif to look for (e.g., AA(N19)TT or AA(N19)NN) [48]. A filter can be enabled in order to avoid stretches of bases of the same kind. Users can also select a species among human, mouse, or rat, as a BLAST parameter for the off-target analysis. Results include the siRNA sense and antisense strands, the distance of the binding sites from start and stop codons, the GC content, the results of the BLAST analysis, and the details about the secondary structure of the siRNA.
The tool DSIR allows the specification of a few prediction parameters as well, such as the siRNA length and the score threshold [49]. Filters for nucleotide stretches and immunostimulatory motifs can also be enabled. Users can choose to design either simple double-stranded siRNAs or shRNA sequences. The tool returns a list of candidate siRNAs (both strands) together with their scores, the complete shRNA sequences (if chosen), and the option to perform a BLAST search on some or all of the predicted siRNAs for the evaluation of off-target effects. Results are exportable in different formats.
siRNA scan is another tool that allows users to specify several design options, other than the length and GC content of the siRNA, such as the 5′ terminal base of the antisense strand, the minimum number of A/U base pairs in seven terminal bases of the antisense strand, and the 5′ terminal base of the sense strand [50]. Stretches of 4 or 9 G/C nucleotides in a row can be avoided, and the number of mismatches in the BLAST similarity search can also be specified.
RNAxs is another tool based on thermodynamics features, concerning local target accessibility in particular [51]. Users can specify design parameters such as the accessibility thresholds, the folding energy, the sequence and energy asymmetry, and the custom sequence constraints. However, default values which have shown to give an optimal separation of functional and nonfunctional siRNAs are already pre-chosen. The output consists of the candidate siRNA sequences, together with accessibility, asymmetry, and self-folding scores. Accessibility plots of the binding sites are also shown, and a BLAST search for similarity can be easily performed by clicking on the provided links.
The tool i-Score, instead, is a sort of “consensus-based tool,” since, in addition to its original score, it also provides nine different designing scores based on different rule sources or other design tools, such as DSIR [52]. For a given target sequence, it returns the complete list of possible siRNAs together with the ten scores, duplex energy, GC content, and the length of the longest GC stretch, highlighting the top ten miRNAs according to i-Score, s-BiopredSi score, and DSIR score.
Finally, we introduce siVirus, a tool for antiviral siRNA design [53]. The system allows the design of siRNA for HIV-1, HCV, SARS, and influenza virus. For each virus, users can select multiple viral subtypes and the target regions in the viral genome. The output consists of a list of siRNA target sites (with the 2-nt overhang), mapped to one or more of the selected genomic regions. For each siRNA, the predicted efficacy according to three different set of rules is given, together with predicted off-target hits and conservation percentage in the selected sequences.
Unfortunately, as mentioned at the end of the previous section, a major issue concerning tools such as the ones described above is represented by the inconsistencies among the current siRNA design rules, mostly due to the heterogeneity of the siRNA data [35]. The Max-Planck Institute devised a principle aimed at identifying all key features relevant to miRNA design. Nevertheless, this effort has shown to yield many noneffective siRNAs which have shown to have a high false-positive rate [54].
A recent meta-rules ensemble strategy which integrates several factors, meta-design rules, and filter criteria has shown to report a 98 % rejection of false-positive miRNAs, showing great improvement over traditional state-of-the-art siRNA design programs [55]. In addition to such strategy, the integration of heterogeneous data sources can greatly alleviate inconsistency issues among siRNA design tools.
3.3 siRNA Databases
Several sources of siRNA sequences are publicly available online. Here we provide a brief overview of some of them, which are listed in Table 3.
Table 3.
siRNA databases
| Database | Data | Link |
|---|---|---|
| NCBI RNAi | siRNA/shRNA sequences | http://www.ncbi.nlm.nih.gov/projects/genome/rnai/ |
| MIT/ICBP siRNA DB | Human and mouse siRNA | http://web.mit.edu/sirna/ |
| HuSiDa | Human siRNA | http://itb.biologie.hu-berlin.de/~nebulus/sirna/ |
| siRecords | Mammalian siRNA | http://sirecords.biolead.org/sirecords |
| VIRsiRNAdb | Antiviral siRNA/shRNA | http://crdd.osdd.net/servers/virsirnadb/ |
For each resource, the kind of data and the link are given
NCBI’ s RNAi resource page allows easy access to the RNAi probes (siRNA/shRNA), stored in the NCBI database. For each probe, details about the sequence, the targets, and the hairpin, in case of an shRNA, are given. Queries are submitted through the standard NCBI interface, which allows results filtering by automatically adding the keywords “gene silencing” to the query.
The MIT/ICBP siRNA Database is a comprehensive database which stores and distributes information on validated siRNAs and shRNAs.
Currently the database contains siRNA and shRNA sequences against over 100 genes from three different sources: (1) sequences designed and tested by MIT researchers, (2) sequences designed by Qiagen and tested by Natasha Caplen’s group at the NCI, and (3) sequences designed by Greg Hannon and Steve Elledge and tested by the ICBP and CGAP programs at the NCI. The database can be searched by keywords (e.g., target gene name) or browsed by gene name and siRNA ID. The results include links to NCBI probe pages. The website also has a section for the submission of new validated reagents. Sequences are available for human and mouse.
HuSiDa is a database that contains sequences of published functional siRNA molecules targeting human genes and important technical details of the corresponding gene silencing experiments [56]. The database is searchable by different terms, such as gene name, cell line, transfection methods, siRNA source, and siRNA sequence.
siRecords archives experimentally tested siRNA inferred from literature [57]. Different data are available for each siRNA, such as its sequence and the alignment with the target gene, the cell types or tissues in which it was tested, the forms of the siRNA agents (e.g., chemically synthesized oligos, vector-transfected shRNA, etc.), and the methods applied to test its efficacy (e.g., Western blot, RT-PCR, etc.). A 4-level efficacy score is assigned to each RNAi experiment based on the data provided by the authors in the original papers. In particular, an experiment is rated as “very high” if the gene product is reduced by more than 90 %, “high” if the gene product is reduced by 70–90 %, “medium” if 50–70 % repression is achieved, and “low” if less than 50 % of the gene product is reduced.
Finally, VIRsiRNAdb is a curated database of experimentally validated viral siRNA and shRNA-targeting genes of 42 human viruses including influenza, SARS, and hepatitis viruses [58]. Currently, the database provides detailed experimental information about 1,358 siRNAs/shRNAs and can be browsed by virus, virus family, gene, and Pubmed ID. It is also searchable by different keywords. For each siRNA, detailed information are shown, including sequence, virus subtype, target gene, GenBank accession, design algorithm, cell type, test object, test method, and efficacy. A section of the database, called EscapeDb, provides information about siRNAs for which viral escape is known, such as the target site mutations.
As previously mentioned, a fundamental issue in siRNA design efficacy and efficiency is represented by the lack of an effective means which would merge all heterogeneous data into the same framework, allowing results based on different data to be comparable.
Different solutions to this nontrivial problem have been devised, but their description is beyond the scope of this chapter. One solution worth mention, though, is the one proposed by Liu et al., which consists in considering each siRNA data source as a “task” and aiming at the development of a joint efficacy model for all the siRNA data sets simultaneously rather than focusing on single data sets in order to derive design rules and efficacy prediction, combining the different results at the end [35].
In conclusion, the main issues with siRNA design are essentially represented by inconsistency among the design rules and improper integration of the cross-platform siRNA data. In a more detailed analysis, aspects such as the lack of a complete feature set and an inadequate consideration of the specificity of target mRNAs could also very well be considered as major causes of the aforementioned problems.
3.4 AntagomiRs: Composition and Resources
A better understanding of the precise molecular function of miRNAs in mammals requires loss-of-function studies in vivo to shed light on a landscape of processes and mechanisms which are to date still largely unknown. To this end, specific and efficient silencers of endogenous miRNAs are necessary tools in aiding research. In 2005, the Krutzfeldt et al. studied the biological significance of silencing miRNAs in vivo with chemically modified, cholesterol-conjugated oligoribonucleotides which they termed “antagomiRs,” disclosing a landscape of therapeutical applications regarding miRNA involved in a disease [32].
The composition of these synthetic RNA analogues basically consists in a hydroxyprolinol-linked cholesterol solid support and 2′-OMe phosphoramidites to make them more resistant to degradation. They essentially reproduce the antisense strand of the endogenous miRNA they inhibit; thus there are no actual design rules applied.
To our knowledge, due to the simple nature of their composition, there are no tools available online for antagomiR design; rather biomedical companies provide researchers with the possibility to chemically synthesize single-stranded, modified RNAs which specifically inhibit endogenous miRNA function after transfection into cells by binding to them and causing the miRNA to be subsequently degraded.
Of relevant importance as a resource for antagomiRs, the database antagomiRBase presents a collection of 53 putative antagomiR sequences for a set of 22 human miRNAs which were used as template in the design of the putative antisense sequences, using GC content and secondary structures of the stem-loop sequences of the miRNAs as parameters, along with the prediction of the free energy of the unbound antagomiRs [59]. The database presents the following information for each miRNA-antagomiR pair: the position of the guide or passenger strand of the miRNA to be targeted in its stem-loop sequence, the actual target and antagomiR sequences, respectively, and the binding energy of the hybrid duplex. A tool is also provided to allow the user to specify a 20–25-nt sequence which is used to query the database, and in case a match in the antagomiR sequences is found, it returns its secondary structure along with all its miRNA targets.
Generally, antagomiRs are efficient as a means to control the expression of specific miRNA molecules. Nevertheless, a valid alternative that could also provide the great advantage of silencing an entire family of miRNAs simultaneously is represented by a group of longer ncRNAs called “sponges,” which we will discuss in the next paragraph.
3.5 Design of Effective miRNA Sponges
Ebert et al. first introduced miRNA sponges in 2007, as an alternative to chemically modified antisense oligonucleotides for miRNA inhibition [34]. Sponges contain multiple binding sites for endogenous miRNAs and function by “absorbing” and distracting them from their natural targets, thus representing a useful tool to probe miRNA functions in a variety of experimental systems.
Sponges can be easily cloned into expression vectors and transiently transfected into cultured cells in order to efficiently derepress miRNA targets. They can also be delivered by virus-based vectors, in order to ensure their stable expression and create continuous miRNA loss of function in cell lines and transgenic organisms [60].
MiRNA binding sites in sponges are usually specific to the miRNA seed region, allowing inhibition of a whole miRNA family. This can represent an advantage over the use of antagomiRs which are highly specific for a single miRNA, being their function dependent upon full complementary match to the miRNA. A single sponge can thus efficiently replace many antagomiRs, with the consequent reduction of potential off-target effects.
Sponges can also be designed to inhibit multiple miRNAs at once. This powerful feature makes them an efficient solution for loss-of-function studies over the traditional knockout model based on miRNA gene deletion, allowing the inhibition of entire genomic and/or functional miRNA clusters, in addition to families. Moreover, the deletion of a single miRNA gene which is part of a cluster could affect the other miRNAs of the cluster, while a sponge represents an efficient and easy way to avoid this side effect and still assure selective inhibition.
Up to date, there are no tools available for the automatic design of single or multiple miRNA sponges; thus we are going to describe some of the design methodologies employed so far in a few successful application.
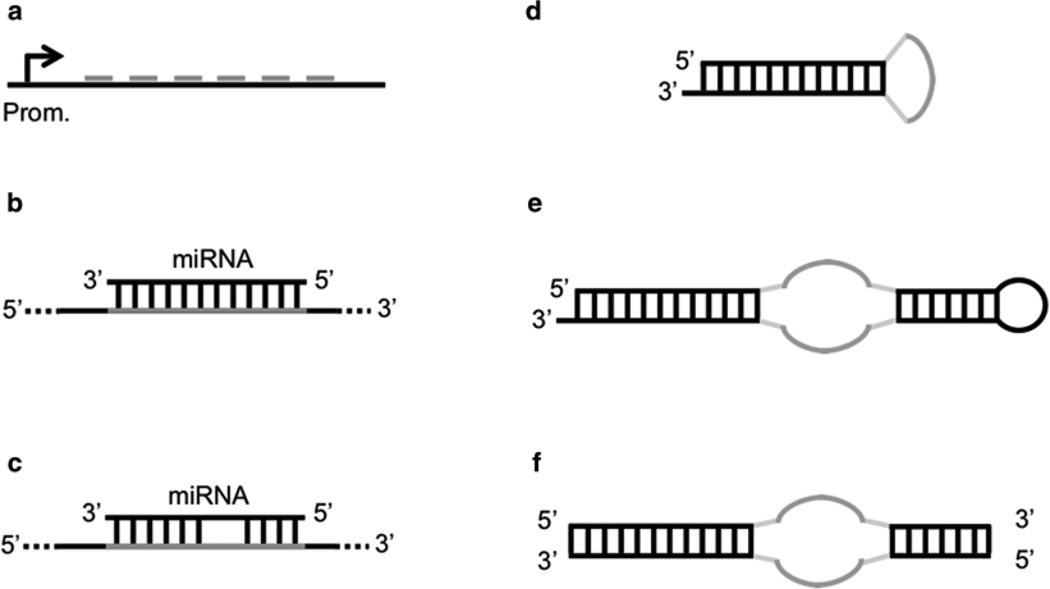
Ebert et al. constructed PolII- and PolIII-generated sponges. PolII sponges were constructed by inserting multiple miRNA binding sites into the 3′ UTR of a destabilized GFP reporter gene driven by the CMV promoter. PolIII sponges were constructed by sub-cloning the miRNA binding region from the GFP construct into a vector containing a U6 snRNA promoter with 5′ and 3′ stem-loop elements. MiRNA binding sites were either bulged or perfectly complementary to the miRNAs. In the first case, a bulge at positions 9–12 of the binding site was introduced in order to prevent cleavage and degradation of the sponge. These sites were separated by 4-nt spacers, while perfect sites had no spacers (Fig. 2b). Both CMV and U6 sponges with 4–7 bulged binding sites produced stronger derepressive effects than sponges with two perfect binding sites. Fluorescence in situ hybridization showed that U6 sponges mainly localized to the nucleus, thus making CMV constructs a better choice. Experiments showed that sponges could selectively inhibit different miRNAs and that a sponge designed for a certain miRNA could also derepress targets of the other miRNAs of the same family. Moreover, the authors suggested 6 as the highest number of functional binding sites, as sponges with more than 6 sites showed a marginal increase in activity above 6 sites. However, they argued that sponges expressed at lower levels could benefit the presence of additional sites.
Fig. 2.
miRNA sponge constructs. (a) Basic sponge with six miRNA binding sites separated by 4-nt spacers. (b) Perfect miRNA binding site on a sponge. (c) Bulged miRNA binding site on a sponge. (d) Prototype decoy consisting of a short hairpin molecule where the loop exposes a binding site for an miRNA. (e) Tough decoy (TuD) with two exposed miRNA binding sites. (f) Synthetic tough decoy (S-TuD) consisting of two fully 2′-O-methylated RNA strands exposing an miRNA binding site each
In subsequent works different types of constructs have been proposed for the expression of miRNA sponges, but from now on we will focus only on the design rules, that being the purpose of this review.
Haraguchi et al. reported optimal conditions for the design of TuD RNAs (tough decoy RNAs), efficient sponges with structurally accessible and indigestible miRNA binding sites [61]. The prototype decoy consisted of a short hairpin molecule where the loop exposed a binding site for an miRNA (Fig. 2c). The length of the stem is critical for the efficient transport of stem-loop structures into the cytoplasm by Exp-5. Experiments determined that the optimal stem length, associated to higher inhibitory effects, was 18 bp. Indeed, stems longer than 18 bp had a reduced binding affinity to Exp-5, while longer stems could be easily digested by Dicer in the cytoplasm. Starting from this prototype, the authors investigated several structural modifications in order to optimize the inhibitory potency of decoys. Experiments showed that the optimal TuD RNA consisted of a bulged stem-loop structure where both sides of the bulge were miRNA binding sites flanked by 3-nt linkers and the two stems separated by the bulge were 18 nt and 8 nt long, respectively (Fig. 2d). The optimal binding sites had a 4-nt insert between nucleotides 10 and 11, in order to avoid cleavage of the decoy. In a subsequent work, the same authors introduced S-TuD (synthetic TuD), a modification of TuD which consists of two fully 2′-O-methylated RNA strands exposing an miRNA binding site each [62]. Following the hybridization of the two paired strands, the resultant S-TuD forms a secondary structure which resembles the corresponding TuD RNA molecule (Fig. 2e). In this work, the authors found that internal base pairing between the two miRNA binding sites on the two strands of a S-TuD can negatively affect the structural accessibility for the miRNA and reduce the inhibitory effect. In light of this, they refined the design rules and suggested that an optimal S-TuD molecule would feature two miRNA binding sites which are perfectly complementary to the target miRNA sequence and which don’t form any base pairing regions longer than 9 nt. If this is not possible, the introduction of a single mutation or a 4-nt insertion in the middle region of the binding sites, as for the TuD previously described, is sufficient in many cases to abolish the base pairing without significantly affecting the affinity to the target miRNA.
These design rules proposed by Haraguchi et al. are the most sophisticated rules described so far for the design of effective miRNA sponges. However, the construction of simple RNA molecules featuring several binding sites for one or more miRNAs, separated by 2–4-nt spacers, is still the most widely used approach. This was confirmed by a recent work of Kluiver et al., in which the authors developed a methodology for the rapid generation of miRNA sponges by making use of simple constructs with up to 20 perfect or bulged miRNA binding sites, as described in the earlier work by Ebert et al. [63].
Nevertheless, it must be noted that despite the optimization of the sponge construct, different application contexts could yield different degrees of inhibition, making the verification of the success of a sponge treatment more challenging than that of genetic miRNA deletion. Thus, it is still under investigation whether in vivo sponge expression can effectively provide a valid alternative to genetic knockouts of miRNA families [60].
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 4.Chu C-Y, Rana TM. Small RNAs: regulators and guardians of the genome. J Cell Physiol. 2007;213:412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- 5.Kutter C, Svoboda P. miRNA, siRNA, piRNA: knowns of the unknown. RNA Biol. 2008;5:181–188. doi: 10.4161/rna.7227. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 8.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 12.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNAs promoters. RNA. 2010;16:495. doi: 10.1261/rna.1731910. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 17.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 18.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 20.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples MicroRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamura K, Phillips MD, Tyler DM, Duan H, Chou Y-T, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunsalus KC, Piano F. RNAi as a tool to study cell biology: building the genomephenome bridge. Curr Opin Cell Biol. 2005;17:3–8. doi: 10.1016/j.ceb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 30.Hadj-Slimane R, Lepelletier Y, Lopez N, Garbay C, Raynaud F. Short interfering RNA (siRNA), a novel therapeutic tool acting on angiogenesis. Biochimie. 2007;89:1234–1244. doi: 10.1016/j.biochi.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 31.de Fougerolles A, Vornlocher H-P, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 33.Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- 34.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Zhou H, Zhu R, Xu Y, Cao Z. Reconsideration of in silico siRNA design from a perspective of heterogeneous data integration: problems and solutions. Brief Bioinform. 2014;15:292. doi: 10.1093/bib/bbs073. doi: 10.1093/bib/bbs073. [DOI] [PubMed] [Google Scholar]
- 36.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strapps WR, Pickering V, Muiru GT, Rice J, Orsborn S, Polisky BA, Sachs A, Bartz SR. The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucleic Acids Res. 2010;38:4788–4797. doi: 10.1093/nar/gkq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 39.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 40.Cui W, Ning J, Naik UP, Duncan MK. OptiRNAi, an RNAi design tool. Comput Methods Programs Biomed. 2004;75:67–73. doi: 10.1016/j.cmpb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 42.Naito Y, Yoshimura J, Morishita S, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinform. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matveeva O, Nechipurenko Y, Rossi L, Moore B, Saetrom P, Ogurtsov AY, Atkins JF, Shabalina SA. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh T, Suzuki T. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 2007;35:e27. doi: 10.1093/nar/gkl1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang P, Wu H, Da Y, Sang F, Wei J, Sun X, Lu Z. RFRCDB-siRNA: improved design of siRNAs by random forest regression model coupled with database searching. Comput Methods Programs Biomed. 2007;87:230–238. doi: 10.1016/j.cmpb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–W108. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schramm G, Ramey R. siRNA design including secondary structure target site prediction. Nat Methods. 2005;2 [Google Scholar]
- 49.Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinform. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 2006;142:429–440. doi: 10.1104/pp.106.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 52.Ichihara M, Murakumo Y, Masuda A, Matsuura T, Asai N, Jijiwa M, Ishida M, Shinmi J, Yatsuya H, Qiao S, et al. Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activities. Nucleic Acids Res. 2007;35:e123. doi: 10.1093/nar/gkm699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naito Y, Ui-Tei K, Nishikawa T, Takebe Y, Saigo K. siVirus: web-based antiviral siRNA design software for highly divergent viral sequences. Nucleic Acids Res. 2006;34:W448–W450. doi: 10.1093/nar/gkl214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong W, Ren Y, Xu Q, Wang Y, Lin D, Zhou H, Li T. Integrated siRNA design based on surveying of features associated with high RNAi effectiveness. BMC Bioinform. 2006;7:516. doi: 10.1186/1471-2105-7-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mysara M, Garibaldi JM, Elhefnawi M. MysiRNA-designer: a workflow for efficient siRNA design. PLoS One. 2011;6:e25642. doi: 10.1371/journal.pone.0025642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Truss M, Swat M, Kielbasa SM, Schäfer R, Herzel H, Hagemeier C. HuSiDa–the human siRNA database: an open-access database for published functional siRNA sequences and technical details of efficient transfer into recipient cells. Nucleic Acids Res. 2005;33:D108–D111. doi: 10.1093/nar/gki131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren Y, Gong W, Zhou H, Wang Y, Xiao F, Li T. siRecords: a database of mammalian RNAi experiments and efficacies. Nucleic Acids Res. 2009;37:D146–D149. doi: 10.1093/nar/gkn817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thakur N, Qureshi A, Kumar M. VIRsiRNAdb: a curated database of experimentally validated viral siRNA/shRNA. Nucleic Acids Res. 2011;40:D230–D236. doi: 10.1093/nar/gkr1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganguli S, Mitra S, Datta A. Antagomirbase: a putative antagomir database. Bioinformation. 2011;7:41. doi: 10.6026/97320630007041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043. doi: 10.1261/rna.2414110. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haraguchi T, Nakano H, Tagawa T, Ohki T, Ueno Y, Yoshida T, Iba H. A potent 2′-O-methylated RNA-based microRNA inhibitor with unique secondary structures. Nucleic Acids Res. 2012;40:e58. doi: 10.1093/nar/gkr1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MKS, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen B-J, van den Berg A. Rapid generation of MicroRNA sponges for MicroRNA inhibition. PLoS One. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsieh A, Bo R, Manola J, Vazquez F, Bare O, Khvorova A, Scaringe S, Sellers W. A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: determinants of gene silencing for use in cell-based screens. Nucleic Acids Res. 2004;32(3):893. doi: 10.1093/nar/gkh238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peek AS. Improving model predictions for RNA interference activities that use support vector machine regression by combining and filtering features. BMC Bioinform. 2007;8:182. doi: 10.1186/1471-2105-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shabalina SA, Spiridonov AN, Ogurtsov AY. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinform. 2006;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 69.Chalk AM, Wahlestedt C, Sonnhammer ELL. Improved and automated prediction of effective siRNA. Biochem Biophys Res Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 70.Klingelhoefer JW, Moutsianas L, Holmes C. Approximate Bayesian feature selection on a large meta-dataset offers novel insights on factors that effect siRNA potency. Bioinformatics. 2009;25:1594–1601. doi: 10.1093/bioinformatics/btp284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Q, Zhou H, Cui J, Cao Z, Xu Y. Reconsideration of in-silico siRNA design based on feature selection: a cross-platform data integration perspective. PLoS One. 2012;7:e37879. doi: 10.1371/journal.pone.0037879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L, Huang C, Yang JY. Predicting siRNA potency with random forests and support vector machines. BMC Genomics. 2010;11:S2. doi: 10.1186/1471-2164-11-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]