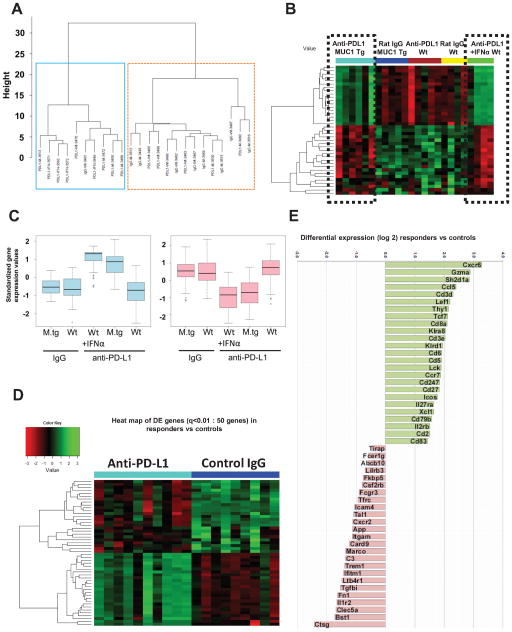
Fig. 5.
Identification of genes associated with anti-PD-L1-induced increase in survival. A Unsupervised cluster analysis using gene expression of n=357 filtered genes, from five mouse groups (n=24): MUC1.Tg mice (n=11, of which 6 received anti-PD-L1 and 5 were controls) and wild type mice (n=13, of which 5 received low dose anti-PD-L1 with no survival benefit, 4 received dose dense anti-PD/L1/IFNα with increased survival and 4 were IgG controls). Two major clusters are observed of 10 (left, blue box) and 14 mice (right, orange box), respectively. B Heat map of the top 39 DE genes, from five mouse group comparisons (ANOVA, p<0.025). Two clusters of genes (of 18 and 21 genes, respectively) show similar patterns in the anti PD-L1 groups with increased survival (black dotted boxes), in contrast to the three other groups with no survival benefit, which also share a similar pattern. C Gene expressions of the two clusters, stratified by groups. Y axis is the standardized expression values for the genes in the cluster. Boxplots show that the anti PD-L1 groups have clearly higher expression than the other groups in the first cluster but lower expression in the second cluster. D Heat map of top 50 DE genes (q< 0.01) from the comparison of anti-PD-L1 treated mice with increased survival (n=10 mice, of which n=6 MUC1.Tg mice treated with anti -PD-L1 and n=4 WT mice treated with dose-intense anti-PD-L1/IFNα) versus isotype control treated mice (non-responders, n=9 mice). E Graph bars of the top 50 DE genes shown in panel D heatmap (q<0.01). Values plotted represent average differential gene expression (log2). Positive values represent upregulated and negative values represent downregulated genes in anti-PD-L1 treated mice compared to controls.