Figure 2.
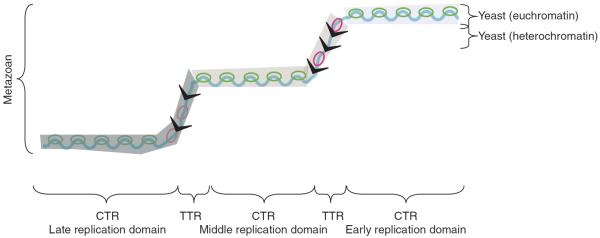
Interpreting replication profiles in different species. A hypothetical segment of a replication timing profile contains regions of constant timing (CTRs), or “replication domains,” that replicate at different times during S phase, and regions of temporal transition (TTRs). However, these regions can be interpreted differently depending on genome size and computational parameters. In metazoan genomes, replication domains can be operationally defined regions where replication timing differs by 10%–20% of the length of S phase. The similarity of replication timing within such domains is proposed to be due in part to the heterogeneous, population-averaged firing of adjacent origins (green bubbles) with similar firing times, but in the case of large CTRs can be due to the nearly simultaneous firing of adjacent but independently regulated replication domains. The actual number of initiation sites within each domain that can potentially be used in a population of cells is believed to be in the dozens (origin clusters) to hundreds (initiation zones). TTRs are regions of suppressed origin activity (indicated as a gray “slime”), which may be replicated either by a single fork (black arrows) moving unidirectionally through time (y-axis) or—if slow-moving forks stimulate firing of inefficient or “dormant” origins—by sequentially activated origins (red bubbles). In contrast, the entire genome in less complex organisms such as yeasts can be thought of as a single replication domain, with the majority of regulation controlled by more origin-proximal mechanisms. The exception is the late-replicating heterochromatin, such as telomeres, which form the equivalents of TTRs, being passively replicated by forks originating in neighboring euchromatic regions.
