Figure 2. Modeling and functional characterization of NOX1 variants.
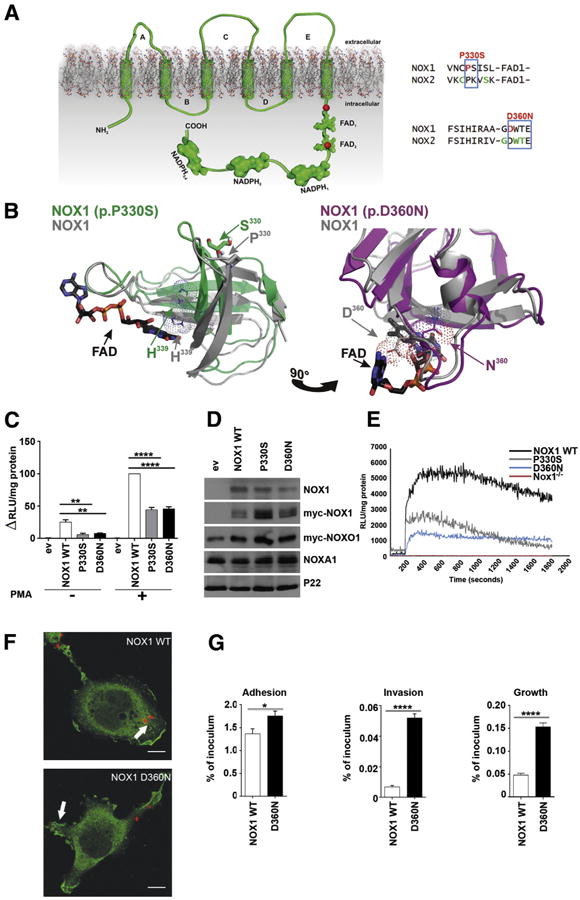
(A) Topologic model depicting NOX1 very early onset inflammatory bowel disease (VEOIBD) variants (red), selected X-CGD CYBB (NOX2) variants (green), conserved residues (blue). (B) Three-dimensional model of NOX1 wild-type (WT) (grey), NOX1 P330S (green), or NOX1 D360N (pink) dehydrogenase domains. NADPH, FAD, residue H339, and variant positions are marked. (C) ROS production by NOX1 WT and variants. (D) Protein expression of NOX1 and variants, Myc-NOXO1, NOXA1, and p22phox as loading control. (E) ROS production in murine Nox1−/− crypts transduced with NOX1 WT or variants. Phorbol 12-myristate 13-acetate (PMA) stimulation was at 200 seconds. (F) Localization of Myc-NOX1 WT or D360N (green) in C. jejuni (red) infected Cos-p22 cells. Scale bar: 10 μm; arrow indicates membrane localization. (G) Adhesion and invasion of Campylobacter jejuni in cells expressing NOX1 WT, P330S, or D360N. Error bars ± standard deviation n = 3; *P ≤ .05; **P ≤ .01; ****P ≤ .0001; comparing NOX1 WT to variants.
