Abstract
Histone deacetylase (HDAC) inhibitors (HDIs) have therapeutic potentials for treating cancer and other diseases. Modulation of gene expression by HDIs is a major mechanism underlying their therapeutic effects. A novel class of HDIs with a previously undescribed benzoylhydrazide scaffold has been discovered through a high throughput screening campaign. Using microarray profiling of gene expression, we have previously demonstrated that treatment of breast cancer cells with a lead benzoylhydrazide HDI UF010 results in cell cycle arrest and apoptosis, likely through activation of tumor suppression pathways with concurrent inhibition of oncogenic pathways. In this brief report, we show methodological and analytical details and discuss additional pathways such as immune signaling that are affected by UF010. Raw and processed data from the microarray were deposited in NCBI’s Gene Expression Omnibus (GEO) database under the accession number: GSE56823.
Keywords: Microarray profiling, Class I histone deacetylase (HDAC) inhibitor, Pathway analysis, Cell cycle, Tumor suppression
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human/MDA-MB-231/Breast, derived from metastatic site (pleural effusion) |
| Sex | Female (51 years adult) |
| Sequencer or array type | Affymetrix GeneChip Human Transcriptome Array 2.0 |
| Data format | CEL files |
| Experimental factors | Cultured MDA-MB-231 cells exposed to DMSO control (n = 3) or benzoylhydrazide HDAC inhibitor UF010 (n = 3) |
| Experimental features | Assess effects of UF010 treatment on global gene expression in cancer cells |
| Consent | N/A |
| Sample source location | ATCC (www.atcc.org) |
Direct link to deposited genomic data
Experimental design, materials and methods
Experimental design
A novel class of small molecule HDIs with a benzoylhydrazide scaffold has been discovered recently [1]. They are specific to class I HDACs 1–3 and appear to exhibit fast-on/slow-off target-binding mechanism. Therefore, the new inhibitors are distinct chemically and mechanistically from known HDIs such as hydroxamic acids and benzamides. As histone deacetylation plays a major role in transcriptional regulation [2–4], we have assessed impact of the new HDIs on global gene expression. We used the triple-negative breast cancer cell line MDA-MB-231 and the benzoylhydrazide analog UF010 to interrogate effects of the new HDIs on gene expression. The experimental design is summarized in Fig. 1.
Fig. 1.
A schematic diagram of gene expression profiling of MDA-MB-231 cells treated with a new HDI (UF010). MDA-MB-231 cells were cultured and then exposed to DMSO (control) or to UF010. Total RNAs were isolated from the treated cells and then processed for hybridization to microarray chips (Affymetrix GeneChip Human Transcriptome Array 2.0). The chips were scanned and data captured. Data were processed (background adjustment, summarization and normalization). The selected statistically significant genes in the experimental groups were analyzed for functional enrichments in certain pathways using Ingenuity Pathway Analysis (IPA) software.
Cell culture and drug treatment
MDA-MB-231 cells were obtained from ATCC and cultured with Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% bovine calf serum, penicillin to 10 units/ml, and streptomycin to 10 μ/ml. Cells (500,000 cells per well) were seeded in a 6-well plate. At 24 h after seeding, dimethyl sulfoxide (DMSO) or UF010 was added. The final concentration for UF010 was 1 μM.
RNA isolation and processing
Total RNAs from the treated cells were isolated using the RNeasy kit (Qiagen) and submitted to the Gene Expression Core of the University of Florida Interdisciplinary Center for Biotechnology Research. A NanoDrop Spectrophotometer (NanoDrop Technologies, Inc.) was used to determine RNA concentration and sample quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.).
Microarray probe preparation, hybridization and data acquisition
The Ambion® WT Expression Kit, the GeneChip WT Terminal Labeling and Controls Kit (Affymetrix) were used for all microarray probe preparation following manufacturer’s protocols. We used 200 ng of total RNA as template for cDNA synthesis. The resulting cDNA was used as template for in vitro transcription (IVT) to generate antisense RNAs, which were then used to produce sense DNA. The sense strand DNA was fragmented, biotinylated, and hybridized with rotation at 45 °C for 16 h to microarray chips (GeneChip® Human Transcriptome Array 2.0, Affymetrix). The arrays were washed and stained with streptavidin-phycoerythrin (SAPE) with an Affymetrix Fluidics Station 450, and scanned using a GeneChip® 7G scanner (Affymetrix).
Microarray data quality control and analysis
The Affymetrix® Expression Console™ Software (Version 1.3) was used to generate.txt files for each RNA hybridization. All subsequent data analyses were performed in R 3.0.0 (http://www.R-project.org/). The Limma package [5] was used for background adjustment, summarization and quantile normalization. Normalization was made using the Robust Multichip Average (RMA) pre-normalization algorithm [6]. Data quality was assessed using various quality control measures. Specifically, density plots were generated to assess log-intensity distributions across a chip. The ideal distributions of the chips show no significant variation (Fig. 2A). An intensity boxplot (Fig. 2B) was used to compare the probe intensity levels between the arrays of the dataset. After normalization, the median lines are not significantly different from each other (Fig. 2B). A heatmap of the six samples was constructed to compare the UF010-treated cells with control (Fig. 2C). For each replicate array, each probe-set signal value from UF010-treated samples was compared to the probe-set signal value of DMSO-treated control samples to give gene expression ratios. Differentially expressed genes were identified using the Limma package with a Benjamini and Hochberg false discovery rate multiple testing correction. The statistically significant or differentially expressed genes were calculated using volcano plot analysis with a fold change (FC) threshold of >1.5 and a p value of <0.05.
Fig. 2.
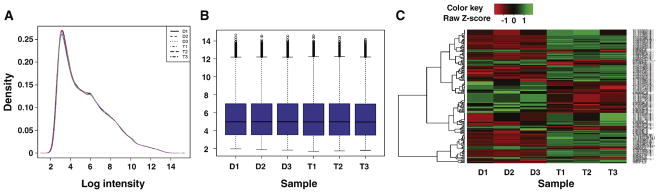
Quality control assessments for the dataset. A, a plot of the log-intensity distribution in the experimental chips of the dataset. B, intensity box plot. The plot compares the probe intensity levels between the arrays of the dataset. Either end of the box represents the upper and lower quartile. The line in the middle of the box represents the median. C, A heatmap comparison of the six samples in the dataset. D, DMSO; T, UF010 treatment.
Ingenuity pathway analysis
The Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems Inc., Redwood City, CA) was used for functional enrichment analysis of the selected statistically significant genes in each of these experimental groups. The association between the genes in the dataset and a functional pathway were made using Fisher’s exact test. Functional groups (or pathways) with a P value less than 0.05 and at least one focused molecule in a pathway were considered to be statistically significant.
Results
Analyses of the microarray dataset reveal that the benzoylhydrazide HDI UF010 induces gene activation and repression, consistent with findings by others that HDIs can up and downregulate gene expression [7–12]. The expression of cell cycle regulators is specifically perturbed by UF010 with downregulation of genes that promote and upregulation of those that inhibit cell-cycle progression [1]. Induction of cell cycle arrest and cell death is a common mechanism of action by HDIs irrespective of cancer or inhibitor types [1,8,9]. Interestingly, UF010 treatment activates major histocompatibility complex (MHC) genes and other genes involved in immune response. Pathway analysis suggests that UF010 specifically induces B-cell receptor, IL-6 and IL-4 signaling. Activation of MHC and immune-related genes by other HDIs in prostate cancer cell lines has been reported [8].
Discussion
The class I HDACs 1–3 play important roles in gene regulation and are overexpressed in cancer [13–17]. HDACs 1 and 2 are closely related and assembled together in deacetylase complexes such as Sin3, NuRD and CoREST [18,19]. HDAC3 is found in the NCOR complex and depends on the deacetylase activation domain (DAD) of NCOR1 or NCOR2 and D-myo-inositol-(1,4,5,6)-tetrakisphosphate for catalyzing deacetylation reaction [20]. Thus, these HDACs may regulate gene expression in a different way and HDIs specific to an individual HDAC isoform may exert distinct impact on gene expression. Different chemical types of small molecule HDIs have been discovered. Hydroxamic acids are potent HDIs and exhibit strong Zn2+-binding property, which limits their isoform selectivity. Nonetheless, isoform selectivity can be achieved through modifying the “cap” moiety that interacts with the residues in the rim outside the substrate tunnel [21]. Benzamide analogs occupy the catalytic center of HDACs and coordinate Zn2+ using the amine and carbonyl groups in a bidentate manner [22]. Benzamide derivatives with a five or six-membered aromatic ring “internal cavity” motif exhibit strong selectivity for HDACs 1 and 2 vs. HDAC3 [22], while HDAC3 selectivity is achieved by modifying the “cap” groups [21]. The benzoylhydrazide HDIs appear to use a linear aliphatic chain to bind the internal hydrophobic cavity of HDACs and they display notable selectivity for HDAC3, while inhibiting HDACs 1 and 2 with similar potency [1]. UF010 shows a fast-on/slow-off target binding mechanism. Whether the unique chemistry and pharmacology of the benzoylhydrazide HDIs can result in distinct modulation of gene expression and therapeutic effects remains to be determined.
Acknowledgments
The authors thank the University of Florida Interdisciplinary Center for Biotechnology Research Gene Expression & Genotype Core (Dr. Yanping Zhang) and Bioinformatics Core (Dr. Jiqiang Yao) for microarray data acquisition and analysis. This work was supported by grants from the Bankhead-Coley Cancer Research Program, Florida Department of Health and the UF Health Cancer Center.
Footnotes
Conflict of interest
None.
References
- 1.Wang Y, Stowe RL, Pinello CE, Tian G, Madoux F, Li D, Zhao LY, Li JL, Wang Y, Wang Y, Ma H, Hodder P, Roush WR, Liao D. Identification of histone deacetylase inhibitors with benzoylhydrazide scaffold that selectively inhibit class I histone deacetylases. Chem Biol. 2015;22:273–284. doi: 10.1016/j.chembiol.2014.12.015. ( http://www.ncbi.nlm.nih.gov/pubmed/25699604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. ( http://www.ncbi.nlm.nih.gov/pubmed/9150133) [DOI] [PubMed] [Google Scholar]
- 3.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. http://www.ncbi.nlm.nih.gov/pubmed/9150137. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. ( http://www.ncbi.nlm.nih.gov/pubmed/19698979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. http://www.ncbi.nlm.nih.gov/pubmed/16646809) [DOI] [PubMed] [Google Scholar]
- 6.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. ( http://www.ncbi.nlm.nih.gov/pubmed/12925520) [DOI] [PubMed] [Google Scholar]
- 7.Kim YJ, Greer CB, Cecchini KR, Harris LN, Tuck DP, Kim TH. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene. 2013;32:2828–2835. doi: 10.1038/onc.2013.32. ( http://www.ncbi.nlm.nih.gov/pubmed/23435418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortenhorst MS, Wissing MD, Rodriguez R, Kachhap SK, Jans JJ, Van der Groep P, Verheul HM, Gupta A, Aiyetan PO, van der Wall E, Carducci MA, Van Diest PJ, Marchionni L. Analysis of the genomic response of human prostate cancer cells to histone deacetylase inhibitors. Epigenetics. 2013;8:907–920. doi: 10.4161/epi.25574. http://www.ncbi.nlm.nih.gov/pubmed/23880963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP, MacKenzie KL, Smyth GK, Johnstone RW. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis. 2013;4:e519. doi: 10.1038/cddis.2013.9. ( http://www.ncbi.nlm.nih.gov/pubmed/23449455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2012;131:777–789. doi: 10.1007/s10549-011-1480-8. ( http://www.ncbi.nlm.nih.gov/pubmed/21452019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, Ding XC, Chanson AL, Reymond MK, Miconnet I, Schrenzel J, Francois P, Calandra T. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. ( http://www.ncbi.nlm.nih.gov/pubmed/20956800) [DOI] [PubMed] [Google Scholar]
- 12.LaBonte MJ, Wilson PM, Fazzone W, Groshen S, Lenz HJ, Ladner RD. DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines. BMC Med Genet. 2009;2:67. doi: 10.1186/1755-8794-2-67. http://www.ncbi.nlm.nih.gov/pubmed/19948057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W, Denkert C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer–overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. ( http://www.ncbi.nlm.nih.gov/pubmed/23627572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weichert W, Denkert C, Noske A, Darb-Esfahani S, Dietel M, Kalloger SE, Huntsman DG, Kobel M. Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia. 2008;10:1021–1027. doi: 10.1593/neo.08474. ( http://www.ncbi.nlm.nih.gov/pubmed/18714364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weichert W, Roske A, Gekeler V, Beckers T, Ebert MP, Pross M, Dietel M, Denkert C, Rocken C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. ( http://www.ncbi.nlm.nih.gov/pubmed/18207460) [DOI] [PubMed] [Google Scholar]
- 16.Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. ( http://www.ncbi.nlm.nih.gov/pubmed/18212746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weichert W, Roske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. ( http://www.ncbi.nlm.nih.gov/pubmed/18347167) [DOI] [PubMed] [Google Scholar]
- 18.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. ( http://www.ncbi.nlm.nih.gov/pubmed/18292778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. ( http://www.ncbi.nlm.nih.gov/pubmed/22414492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. ( http://www.ncbi.nlm.nih.gov/pubmed/22230954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner FF, Weiwer M, Lewis MC, Holson EB. Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics. 2013;10:589–604. doi: 10.1007/s13311-013-0226-1. ( http://www.ncbi.nlm.nih.gov/pubmed/24101253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauffer BE, Mintzer R, Fong R, Mukund S, Tam C, Zilberleyb I, Flicke B, Ritscher A, Fedorowicz G, Vallero R, Ortwine DF, Gunzner J, Modrusan Z, Neumann L, Koth CM, Lupardus PJ, Kaminker JS, Heise CE, Steiner P. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem. 2013;288:26926–26943. doi: 10.1074/jbc.M113.490706. ( http://www.ncbi.nlm.nih.gov/pubmed/23897821) [DOI] [PMC free article] [PubMed] [Google Scholar]