Abstract
Circadian oscillators are posttranslationally regulated and affect gene expression in autotrophic cyanobacteria. Oscillations are controlled by phosphorylation of the KaiC protein, which is modulated by the KaiA and KaiB proteins. However, it remains unclear how time information is transmitted to transcriptional output. We show reconstruction of the KaiABC oscillator in the noncircadian bacterium Escherichia coli. This orthogonal system shows circadian oscillations in KaiC phosphorylation and in a synthetic transcriptional reporter. Coexpression of KaiABC with additional native cyanobacterial components demonstrates a minimally sufficient set of proteins for transcriptional output from a native cyanobacterial promoter in E. coli. Together, these results demonstrate that a circadian oscillator is transplantable to a heterologous organism for reductive study as well as wide-ranging applications.
Circadian oscillators regulate metabolic and behavioral changes in anticipation of day-night variations in environmental conditions and are shared by organisms across the tree of life (1). In the cyanobacterium Synechococcus elongatus, a well-studied system for circadian rhythm regulation, changes in the phosphorylation state of KaiC are coordinated with diurnal cycles. During the day, KaiA promotes the autokinase activity of KaiC, resulting in phosphorylated KaiC at subjective dusk. At night, KaiB antagonizes the activity of KaiA, promoting the autophosphatase activity of KaiC and returning KaiC to the unphosphorylated state again at subjective dawn (Fig. 1A) (2, 3). These changes drive corresponding oscillations in gene expression. The KaiABC system has been reconstituted in vitro, and phosphorylation of KaiC oscillates in this minimal system (4). These findings suggest that the cyanobacterial oscillator could function when heterologously expressed in other organisms, which would have practical applications as well as implications for the understanding of oscillatory behavior evolution. Here, we demonstrate functionality of the reconstructed Kai circadian oscillator in Escherichia coli, a gut bacterium without its own native circadian rhythm.
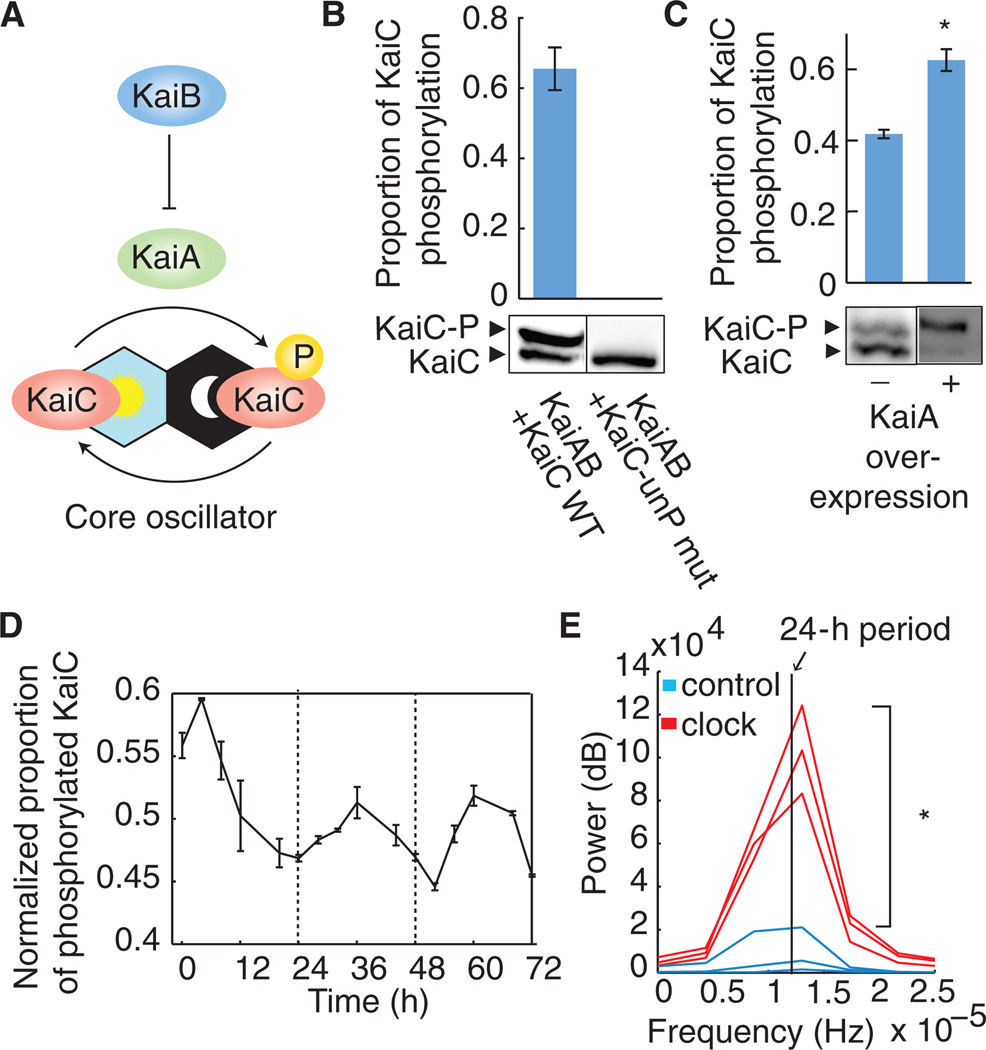
Fig. 1. Functionality of the core KaiABC oscillator in a heterologous system.
(A) Model for the phosphorylation state changes of KaiC through the antagonistic activities of KaiA and KaiB. (B) Levels of phosphorylated (−P) and unphosphorylated wild-type (WT) or nonphosphorylatable mutant (S431A, T432A; “KaiC-unP mut”) of KaiC-FLAG protein when expressed with KaiAB. Proteins were visualized by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting (see Materials and Methods). The ratio of phosphorylated KaiC to total KaiC is plotted. (C) Levels of phosphorylated and unphosphorylated KaiC-FLAG, coexpressed with KaiB, with or without KaiA overexpression (see Materials and Methods). The ratio of phosphorylated KaiC to total KaiC is plotted. The asterisk indicates higher proportion of phosphorylated KaiC when KaiA is overexpressed (P < 0.05, Student’s t test). (D) KaiC phosphorylation over time in E. coli coexpressing KaiA and KaiB, after synchronization (t = 0 hours). The mean ratio of phosphorylated KaiC to total KaiC across biological replicates, mean-normalized for each time trace, is plotted. Circadian oscillations are statistically significant as analyzed using RAIN (P < 0.001) (7). (E) Power of circadian periodicity of KaiABC clock strains (red) and control strains expressing only KaiC (blue). Traces produced by bandpass filtering for circadian periods (20 to 30 hours) followed by Fourier transform of time course data from biological replicates are plotted (n = 3). Vertical line indicates frequency corresponding to a 24-hour period. The asterisk indicates increased power of circadian periodicity in KaiABC clock strains (P < 0.05, Student’s t test). Error bars, SEM (n = 3) (B to D).
When KaiABC is expressed in E. coli, KaiC phosphorylation is regulated by KaiA and KaiB. To reconstruct the oscillator, we expressed FLAG-tagged KaiC in an operon with KaiA and KaiB in E. coli (fig. S1A). Phos-tag Western blot was used to visualize KaiC-FLAG (Fig. 1, B to D, and fig. S2). Two protein bands at ~60 kD, corresponding to the size of KaiC monomers, were observed (Fig. 1B). The upper band was phosphorylated KaiC (KaiC-P) because it did not appear in a nonphosphorylatable mutant (S431A, T432A) of KaiC (5) (Fig. 1B). Moreover, overexpression of KaiA resulted in a higher proportion of phosphorylated KaiC (Fig. 1C; P < 0.05, Student’s t test) as would be expected from the known function of KaiA (6) (Fig. 1A). We expressed kaiA and kaiBC under two distinct inducible promoters and observed that phosphorylated KaiC dominates (63%) in strains with high expression of KaiA and unphosphorylated KaiC dominates (42%) in strains with no KaiA expression (Fig. 1C).
KaiC phosphorylation over time showed changes consistent with a circadian rhythm. We assayed KaiC phosphorylation (KaiC-P) by Western blot in a strain expressing KaiABC and quantified the ratio of phosphorylated KaiC to total KaiC by densitometry. After adenosine triphosphate (ATP) deprivation by transient minimal medium shock (at t = 0) to synchronize KaiC-P levels, we observed oscillations in KaiC-P levels with a circadian period of about 24 hours for 3 days (Fig. 1D and fig. S2A). Phosphorylation levels over time were analyzed using RAIN (7), and circadian oscillations were shown to be statistically significant (P < 0.001). The proportion of unphosphorylated KaiC oscillated as well, but in antiphase as expected (fig. S2B). Total KaiC expression showed no oscillating trends over time (RAIN, P = 0.35) (fig. S2C), and control strains expressing only KaiC without KaiA and KaiB showed increases in KaiC-P levels over time but also showed no circadian trends (fig. S3; RAIN, P > 0.99). Moreover, we compared the strength of circadian periodicity at 24 hours between KaiABC clock strains and KaiC alone control strains (Fig. 1E). To do so, we first conducted background subtraction of other periodicities outside of the circadian range (20 to 30 hours) (8) by bandpass filtering. We then performed Fourier analysis, which allowed us to quantify the power of each trace at the circadian period (24 hours) and make direct comparisons between Kai clock and control strains. We found that the mean power at the 24-hour period of KaiABC clock strains was significantly higher than that of KaiC alone control strains (Fig. 1E; P < 0.05, Student’s t test). These results indicated that KaiC phosphorylation oscillations, which required the presence of KaiA and KaiB, were circadian with a period of ~24 hours.
In order to affect cellular physiology, a posttranslational circadian clock needs to be connected to transcriptional output. To do so, we built a synthetic oscillator utilizing a modified bacterial two-hybrid system (9, 10) (Fig. 2A). Phosphorylated KaiC has been shown to bind more strongly than unphosphorylated KaiC to SasA, a downstream component to the core KaiABC oscillator (11) (Fig. 2A, inset). This property allowed us to build a synthetic transcriptional reporter of circadian state consisting of two binding partners: (i) full-length KaiC C-terminally fused to the α subunit N-terminal domain of RNA polymerase (KaiC-αNTD) and (ii) full-length SasA C-terminally fused to λCI protein (SasA-CI) (Fig. 2A and fig. S4). Only when the two fusion proteins interact can they then bind to the promoter and activate transcription of gfp.
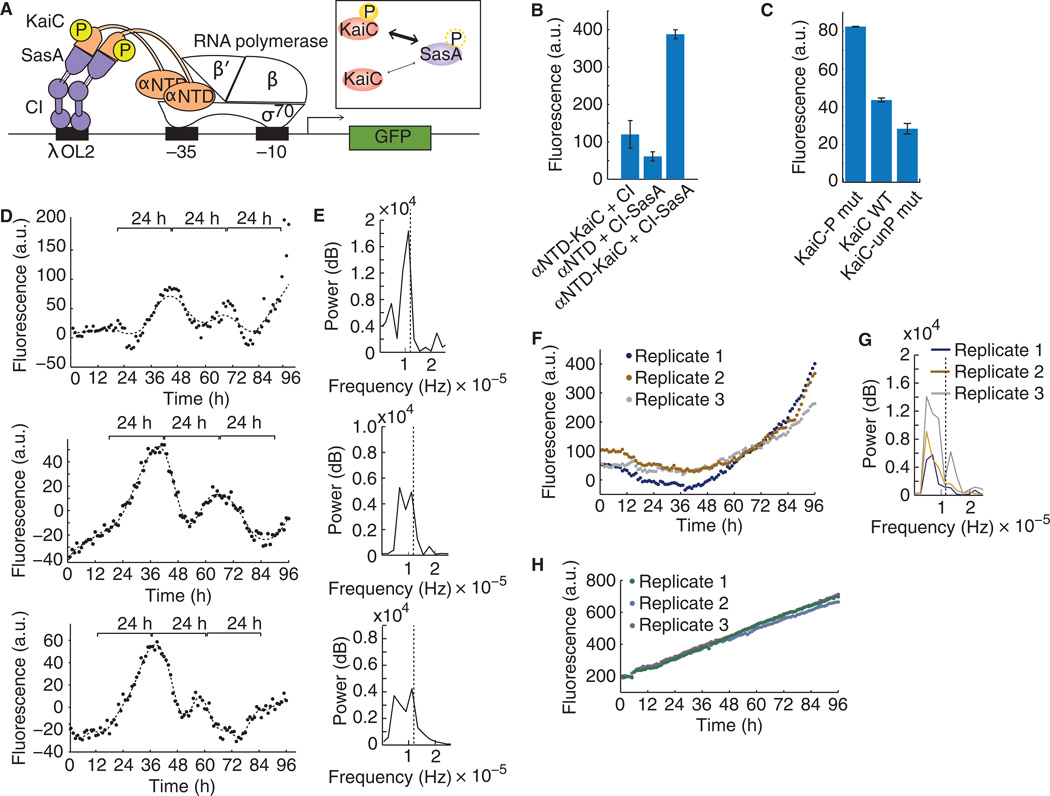
Fig. 2. Asynthetic transcriptional reporter of KaiC phosphorylation demonstrates circadian oscillations.
(A) Schematic of the synthetic transcriptional reporter of KaiC phosphorylation state. Expression of the green fluorescent protein (GFP) reporter is dependent on the interaction between KaiC, which is fused to the α subunit of RNA polymerase N-terminal-domain (KaiC-αNTD), and SasA, which is fused to λCI binding protein (SasA-CI). Inset: Thickness of arrows indicates differing binding strength of phosphorylated and unphosphorylated KaiC to SasA. (B) OD-normalized fluorescent reporter output from strains with either binding partner alone or both KaiC-αNTD and SasA-CI. Error bars, SEM (n = 3). (C) OD-normalized fluorescent reporter output of interactions between phosphomimic KaiC (S431A, T432E; −Pmut), wild-type KaiC (WT), or a nonphosphorylatable mutant of KaiC (S431A, T432A; −unPmut) with WT SasA-CI. Error bars, SEM (n = 3). (D to G) Background-subtracted and OD-normalized time course fluorescence when coexpressing KaiC (D) or KaiC S431A T432A nonphosphorylatable mutant (F) with SasA fusion proteins and KaiAB. Brackets indicate 24-hour periods. Statistically significant circadian oscillations were observed in (D) using RAIN (P < 0.05). Maximum power, shown by Fourier analysis of (D) and (F), occurs at a period of ~24 hours in (E) and at periods of >30 and/or <20 hours in controls (G). Dashed line indicates a 24-hour period. t = 0 indicates synchronization. (H) Background fluorescence, which was subtracted in (D) and (F), from a strain containing only the reporter and no clock components.
Coexpression of the two binding partners, SasA-CI and KaiC-αNTD, with the remaining components of the core KaiABC oscillator, KaiA and KaiB, resulted in transcriptional outputs dependent on the presence of both binding partners as well as the phosphorylation state of KaiC. When compared to controls containing only KaiC-αNTD or only SasA-CI, gene expression from the synthetic reporter containing both fusion proteins was about threefold higher after overnight induction (Fig. 2B), indicating that both binding partners are necessary for reporter activation. A phosphomimic of KaiC showed higher reporter output compared to nonphosphorylatable KaiC, indicating that the KaiC phosphorylation state determines reporter activity (Fig. 2C). We also observed that changes in SasA phosphorylation could affect reporter output (fig. S5), but these changes were not necessary; KaiC phosphorylation changes alone were sufficient to change reporter output. These results (Fig. 2, B and C), together with our previous observation that KaiC phosphorylation state shows circadian oscillations (Fig. 1D), suggest that KaiC circadian oscillations can be transmitted to a transcriptional output using this synthetic reporter.
Indeed, the transcriptional reporter showed circadian behavior over time; the synthetic circadian transcriptional system oscillated with a ~24-hour period (RAIN, P < 0.05, n = 3) for more than 3 days after minimal medium synchronization (Fig. 2D and table S1). All biological replicates showed this trend, although there was some variability in phase and amplitude, likely due to a lack of redundant mechanisms, such as those found in cyanobacteria (12, 13), to maintain robustness and synchronization. Background reporter fluorescence alone showed linear increases over time without any circadian periods (Fig. 2H). Fourier analysis revealed a dominant frequency with a period between 20 and 30 hours (Fig. 2E). A negative control, consisting of a nonphosphorylatable mutant of KaiC with all other components remaining the same, showed fluctuations in fluorescence but no dominant period (or peak in the Fourier spectra) at 20 to 30 hours (Fig. 2, F and G).
Consistent with the observed population-wide circadian oscillations, we found that oscillations can also be observed at the single-cell level. Cells were loaded in channels of a microfluidic device that only retains and analyzes a single dividing “mother” cell and not its progeny (14, 15) (Fig. 3, A and B, and movie S1). This device is open on one end to the medium flowing across the bottom, perpendicular to the channel (Fig. 3A). Daughter cells were washed away, where as mother cells were tracked across multiple cell divisions (Fig. 3A). Single-cell fluorescence time trace data (Fig. 3, B and C) were collected for both circadian circuit–containing cells and the corresponding control strains containing reporter alone. As above, to examine the frequencies of interest, we applied a bandpass filter to the data from both Kai cock and control strains, only allowing for signals from circadian periodicities of 20 to 30 hours (8) (Fig. 3D). Fourier analysis provided a quantitative measurement of the strength at each frequency (power), which allowed us to make direct comparisons between oscillation periods in Kai clock and control strains. Kai clock strains showed an enrichment of high-power signals with ~24-hour periods compared to controls, which still show a small amount of power near this period, likely due to cellular noise (Fig. 3E). In addition, the single-cell oscillations are sensitive to the starting conditions for each cell. However, the overall distribution of power of the circadian clock cells was greater than that of controls (one-tailed, two-sample Kolmogorov-Smirnov test, P = 0.001) (Fig. 3E), indicating that circadian oscillations at the population level are likely a result of single-cell oscillations.
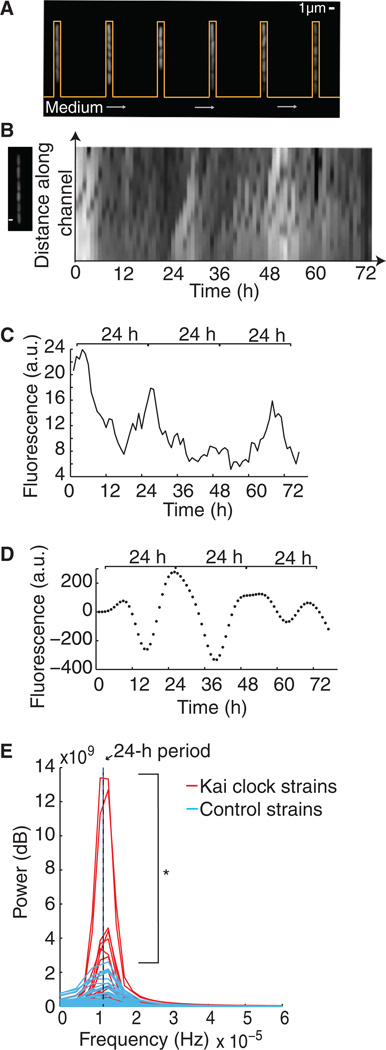
Fig. 3. Single cells demonstrate oscillatory behavior.
(A) Fluorescent cells in channels in the microfluidic device (orange outline) with medium flowing (gray arrows) across the base of the channels, allowing for long-term microscopy assay of single-cell fluorescence. (B) Fluorescent cells in one channel of the microfluidic device at one time point (left). Kymograph shows fluorescence of a single channel over time (right), after overnight induction and minimal medium shock synchronization (at t = 0). Time interval, 1 hour. (C) Average fluorescence of a single mother cell, grown as in (A) and (B), containing the synthetic reporter and Kai clock components. (D) Bandpass-filtered (circadian periods of 20 to 30 hours) data from (B) to compare the strength of circadian periodicity across multiple single-cell traces. (E) Fourier spectra of the time courses of Kai clock strains (red, N = 65) or control strains containing only the reporter (blue, N = 48), both filtered as in (D). Eleven percent of cells had a higher power than the control cell with the highest power. Asterisk indicates that the overall distribution of power of the circadian clock cells is greater than that of controls (one-tailed, two-sample Kolmogorov-Smirnov test, P = 0.001). Black line indicates frequency corresponding to a 24-hour period.
A posttranslational oscillation signal can be transmitted to a natural output by introducing additional Kai clock components and circadian-responsive promoters from cyanobacteria into E. coli together with the basic KaiABC system. Several proteins including RpaA, SasA, and RpaB have been implicated in the transmission of the Kai signal (16–19). We constructed E. coli strains (denoted “reporter + Kai clock”) containing kaiABC in an operon driven by an arabinose-inducible promoter; a synthetic operon containing components sasA, rpaA, and rpaB driven by a lac promoter; and a fluorescent protein reporter driven by one of three circadian-responsive promoters (18) from S. elongatus: kaiBC, rpoD6, or sigF2 (fig. S1). Overnight induction in unsynchronized cells resulted in reporter expression using each of the three promoters (Fig. 4A). The kaiBC promoter (PkaiBC) showed the highest fold change (7.6) between the reporter + Kai clock strain and a strain with only the reporter. In contrast, the same reporter driven by the promoter of an E. coli housekeeping gene, ihfB, showed decreased expression when coexpressed with the Kai clock and downstream components (Fig. 4B), likely due to cellular burden when expressing heterologous components. These results indicate that the Kai components are driving reporter expression from cyanobacterial promoters in the heterologous system.
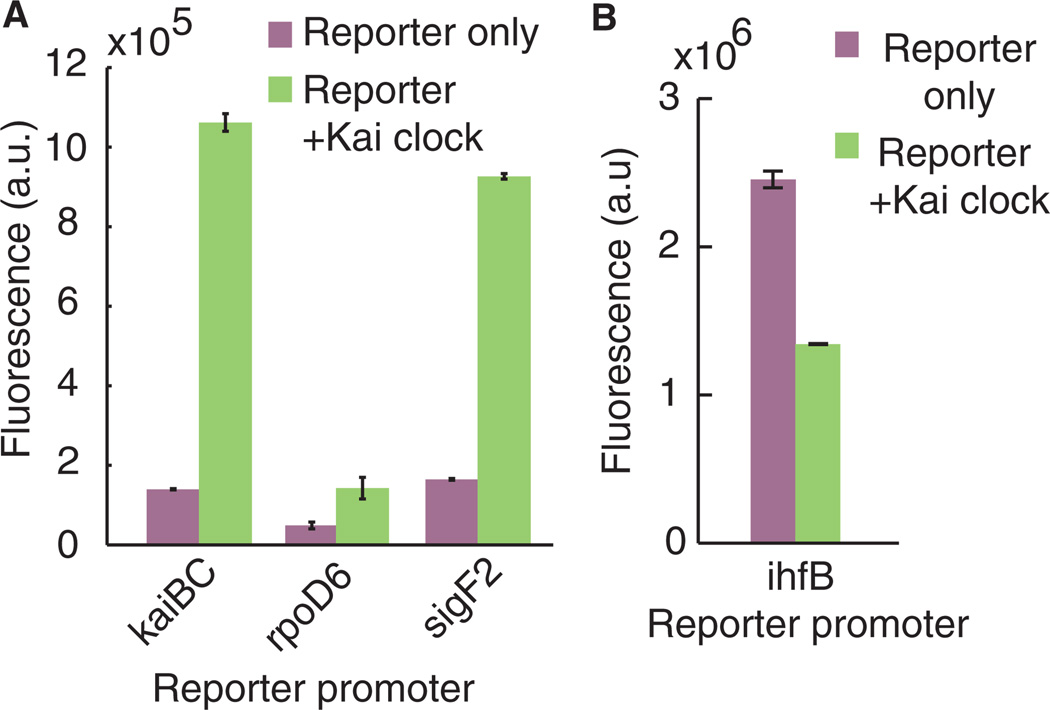
Fig. 4. KaiC transmits phosphorylation information to cyanobacterial circadian promoters in E. coli.
(A) Reporter fluorescence driven by cyanobacterial circadian-responsive promoters (kaiBC, rpoD6, or sigF2), with or without the Kai clock components. (B) Reporter fluorescence driven by E. coli housekeeping gene promoter, ihfB, with or without Kai clock components. All measurements were taken after overnight induction. Error bars, SEM (n = 3).
Our system demonstrates the transplantability of the bacterial circadian oscillator and its potential for reengineering. Whereas existing devices can program time delays (20) and produce cellular oscillators with relatively short periods (21), our oscillator has a period that matches the geophysical day-night cycle. We thereby improve the reach of temporal circuits in synthetic biology to encompass a biologically relevant periodicity. Recently, it has been shown that the disturbance of gut microbiota circadian rhythms, which are dictated by feeding rhythms, can result in host metabolic imbalances, such as obesity and glucose intolerance (22). Modulation of gut microbes’ circadian rhythms through engineering may therefore be a plausible strategy for the correction of jet lag–induced dysbiosis. More broadly, our system will be pertinent as a platform for a variety of biotechnological applications, such as automated daily drug delivery, circadian control in industrial microbial processes, and treatment of circadian rhythm disorders (23, 24).
MATERIALS AND METHODS
Strains, plasmids, and growth conditions
The relevant plasmids and strains are presented in table S2. All plasmids were cloned using Gibson assembly (25) or restriction cloning. All plasmids were verified by sequencing. Phosphomutants were constructed by mutating the phosphorylated residue to a glutamic acid to mimic the constitutively phosphorylated state, or to an alanine to make it nonphosphorylatable. All phosphomutants were constructed using Q5 Site-Directed Mutagenesis kits following the manufacturer’s instructions (New England Biolabs). E. coli Top10 strain (Life Technologies) was used in all experiments with two exceptions. For constructs with arabinose-inducible promoters, the strain used was DP10, a previously described TOP10 variant with regulatable control of pBAD (26). MG1655 E. coli strain was used in single-cell experiments for ease of loading and growth in the microfluidic chamber.
For KaiA overexpression, kaiA was driven by a pBAD promoter and kaiBC was driven by pTET [aTc (400 ng/ml) induction]. Induction with 500 µM arabinose was used for overexpression of KaiA versus 0 µM arabinose induction for no overexpression. KaiC phosphorylation reporter strains contained a synthetic interaction-dependent reporter, KaiC-α N-terminal domain (αNTD), and SasA-λCI. To avoid SasA being trapped in the phosphorylated state, RpaA and CikA, components involved in the dephosphorylation of SasA, were expressed for all time course experiments.
Unless otherwise stated, cells were grown in LB medium with appropriate antibiotics overnight; single colonies were picked intoM9 minimal medium with 0.4% glycerol, 0.1% casamino acids, antibiotics, and 500 µM arabinose induction and/or 20 µM isopropyl-β-d-thiogalactopyranoside induction, as necessary. All end-point measurements were taken after overnight growth in induction medium.
Phos-tag SDS-PAGE and Western blot
Cell lysates were prepared by incubation in 3% SDS for 10 min at 100°C. Prepared lysates were separated by SuperSep Phos-tag precast gels following the manufacturer’s protocol (Wako Chemicals). After four washes (10 min each) in transfer buffer (0.025 M tris base, 0.192 M glycine, 10% methanol) with 10 mM EDTA and one additional wash in transfer buffer without EDTA, proteins were transferred onto polyvinylidene difluoride membranes using an iBlot Gel Transfer Device and iBlot Gel Transfer Stacks (Life Technologies). After overnight blocking in 10% bovine serum albumin to reduce background binding, the membranes were blotted using the SNAP-ID Protein Detection System (EMD Millipore) following the manufacturer’s instructions. Proteins of interest were probed with anti-FLAG (Abcam, ab1238) or anti–6xHis tag (Abcam, ab1187) antibodies, both used at a final dilution of 1:10,000. Peroxidase conjugates were detected using SuperSignal West Dura Extended Duration substrate (Thermo Scientific). Quantification was performed in FIJI (27) using densitometry. The proportion of KaiC phosphorylation was determined by normalizing the intensity of the upper band (phosphorylated KaiC) to the sum of the intensities of both the upper and lower bands (total KaiC).
Fourier analysis
Discrete Fourier transform was performed using a built-in MATLAB function. Unless otherwise stated, Butterworth bandpass filters were applied as previously described (28) to subtract background periodicities outside of the range of interest. The low-pass filter was set at the measurement interval (1 hour) and the high-pass filter was set at the acquisition time of the experiment to enable determination of the period. When directly comparing the strength of circadian periodicity at 24 hours between control strains and clock strains, background subtraction of other periodicities outside of the circadian range (20 to 30 hours) was conducted by bandpass filtering.
Time course measurements
Cells grown as described above were transferred to M9 minimal medium with no carbon source for 1 hour to synchronize the population (29). Cells were back diluted to an OD600(optical density at 600 nm) of ~0.1 in either 500 ml (for KaiC phosphorylation analysis) or 200 µl of M9 minimal medium (for plate reader analysis) with 0.5% succinate and 1 mM leucine to promote slower growth and without induction to reduce population desynchronization. For analysis of KaiC phosphorylation, aliquots were taken at 4-hour intervals and lysed by incubation in 3% SDS for 10 min at 100°C and visualized on a Western blot as described above. Mean normalization was performed on each time course trace from the three biological replicates. The mean-normalized, average proportion of KaiC phosphorylation was plotted. For fluorescence time courses of the synthetic reporter, mineral oil was added to prevent evaporation. Readings were taken using a Synergy NEO HTS MultiMode microplate reader (BioTek) using appropriate filters. Fluorescence readings were normalized to OD600, and background (OD600 normalized fluorescence readings from a reporter only strain AHC157) was subtracted. For both KaiC phosphorylation and synthetic reporter time courses, statistical analyses were performed with RAIN (7) using a periodicity of 24.8 hours over a 3-day time course.
Imaging cells in a microfluidic device
Cells were grown as described above to log phase (OD600 ~0.3) and then resuspended in M9 minimal medium with 0.5% succinate, 1 mM leucine, and Pluronic F107 (0.85 g/liter), a surfactant to aid loading. The microfluidic chip was prepared, loaded, and imaged as previously described (15). Image acquisition was performed once per hour using a custom MATLAB script and µManager (30) as described previously (15).
Single-cell imaging data analysis
Cell outlines were automatically determined using MicrobeTracker (31). The average cell fluorescence of the mother cell (the bottom-most cell of each channel) was tracked over time. All subsequent analysis was performed using MATLAB. To examine frequencies of interest and to directly compare the strength of circadian periodicity between the experimental and control samples, Butterworth bandpass filters were applied to traces as previously described (28), with the boundaries set at 20 to 30 hours, which are by definition circadian periodicities (8). Discrete Fourier transforms were performed, and power was determined. The frequency corresponding to a 24-hour period was calculated to be 1.15 × 10−5 Hz. Power at that frequency was interpolated from the Fourier spectra. A two-sample, one-sided Kolmogorov-Smirnov test was performed, comparing the population of cells with circadian clocks with controls cells lacking the clock.
End-point measurements
Cells were grown as described above. Fluorescence and OD600 were measured in triplicate on a Victor 3V 1420 Multilabel Counter (Perkin-Elmer) using appropriate filters. Fluorescence was normalized to OD. Error bars are SEM.
Supplementary Material
Acknowledgments
We thank N. Lord and J. Paulsson for advice and assistance on single-cell experiments, as well as P. Deighan and A. Hochschild for assistance on bacterial two-hybrid experiments. Thanks to J. Way, S. Hays, T. Ford, and J. Torella for comments and discussions. We would also like to thank the Harvard Medical School microfluidics core facility. Funding: This work was supported by Defense Advanced Research Projects Agency grant N66001-11-C-4203. A.H.C. was supported by NIH grant GM080177 and an NSF Graduate Research Fellowship. R.L.C. was supported by Gordon and Betty Moore Foundation postdoctoral fellowship through the Life Sciences Research Foundation (GBMF 2550.04).
Footnotes
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/5/e1500358/DC1
Fig. S1. Plasmids built for reconstruction of the circadian oscillator in E. coli.
Fig. S2. Additional data and quantifications of KaiC phosphorylation in E. coli expressing KaiABC.
Fig. S3. Circadian phosphorylation of KaiC over time requires KaiA and KaiB.
Fig. S4. Plasmids built for the synthetic oscillator utilizing a modified bacterial two-hybrid system.
Fig. S5. KaiC and SasA phosphorylation states affect reporter output.
Table S1. Raw data for Fig. 2D.
Table S2. Bacterial strains and plasmids.
Movie S1. Circadian oscillations visualized in single E. coli cells using a microfluidic device.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y-I, Dong G, Carruthers CW, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 5.Ito H, Kageyama H, Mutsuda M, Nakajima M, Oyama T, Kondo T. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat. Struct. Mol. Biol. 2007;14:1084–1088. doi: 10.1038/nsmb1312. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J. Biol. Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 9.Dove SL, Hochschild A. A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 2004;261:231–246. doi: 10.1385/1-59259-762-9:231. [DOI] [PubMed] [Google Scholar]
- 10.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein–protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 11.Valencia S J, Bitou K, Ishii K, Murakami R, Morishita M, Onai K, Furukawa Y, Imada K, Namba K, Ishiura M. Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells. 2012;17:398–419. doi: 10.1111/j.1365-2443.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- 12.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwicker D, Lubensky DK, ten Wolde PR. Robust circadian clocks from coupled protein-modification and transcription–translation cycles. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22540–22545. doi: 10.1073/pnas.1007613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, Jun S. Robust growth of Escherichia coli. Curr. Biol. 2010;20:1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norman TM, Lord ND, Paulsson J, Losick R. Memory and modularity in cell-fate decision making. Nature. 2013;503:481–486. doi: 10.1038/nature12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markson JS, Piechura JR, Puszynska AM, O’Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanaoka M, Takai N, Hosokawa N, Fujiwara M, Akimoto Y, Kobori N, Iwasaki H, Kondo T, Tanaka K. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J. Biol. Chem. 2012;287:26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol. Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber W, Stelling J, Rimann M, Keller B, Daoud-El BM, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 22.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 23.Wirz-Justice A, Benedetti F, Berger M, Lam RW, Martiny K, Terman M, Wu JC. Chronotherapeutics (light and wake therapy) in affective disorders. Psychol. Med. 2005;35:939–944. doi: 10.1017/s003329170500437x. [DOI] [PubMed] [Google Scholar]
- 24.Lévi F. From circadian rhythms to cancer chronotherapeutics. Chronobiol. Int. 2002;19:1–19. doi: 10.1081/cbi-120002676. [DOI] [PubMed] [Google Scholar]
- 25.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 26.Kizer L, Pitera DJ, Pfleger BF, Keasling JD. Application of functional genomics to pathway optimization for increased isoprenoid production. Appl. Environ. Microbiol. 2008;74:3229–3241. doi: 10.1128/AEM.02750-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine J, Funes P, Dowse H, Hall J. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman AG, Fall L, Atkinson DE. Adenylate energy charge in Escherichia coli during growth and starvation. J. Bacteriol. 1971;108:1072–1086. doi: 10.1128/jb.108.3.1072-1086.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using µManager software. J. Biol. Methods. 2014;1:10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.