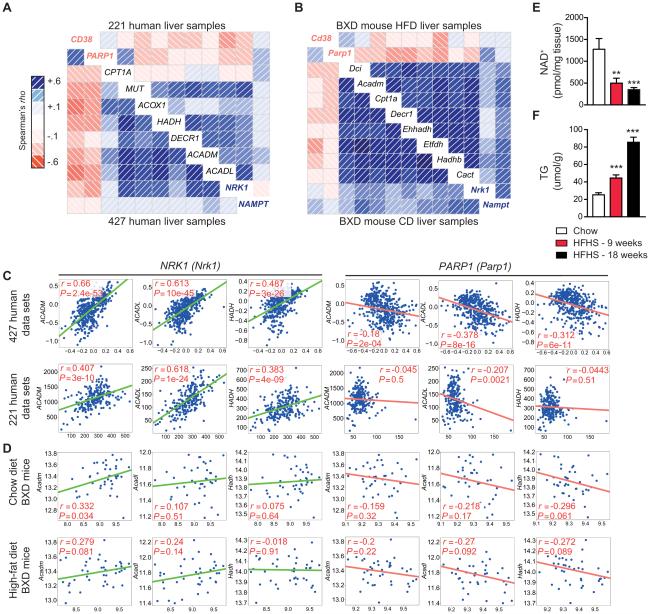
Fig. 1.
NAD+ regulating enzymes and NAD+ levels correlate with lipid metabolism in both humans and mice. Transcripts from NAD+-synthesis genes (shown in blue font), in contrast to NAD+-consuming genes (shown in red font), were positively correlated with regulators of β-oxidation using a custom designed data-set derived from (A) two human data sets, including 427 (28) and 220 (27) human liver samples or (B) two mouse data sets, including 42 BXD strains fed either chow or high-fat diets (9, 26). As seen on a correlogram, blue correlations are positive (red correlations are negative – intensity of the colors correlates with level of significance). NRK1 and NAMPT transcripts, in contrast to Parp1, were positively correlated to mitochondrial β-oxidation genes, ACADL, ACADM and HADH, in data sets from (A, C) 427 (28) and 220 (27) human liver samples or 42 BXD strains fed either (B, D) chow or high-fat diets (9, 26) Consistent with these findings in humans, we see reductions in hepatic (E) NAD+ levels, followed by increases in (F) hepatic triglyceride levels, in mice fed 9- to 18-weeks with a HFHS diet (n=5-10). *P < 0.05, **P < 0.001, ***P < 0.0001 compared to the CD cohort. Data are expressed as mean ± s.e.m. One-way ANOVA with a post-hoc Bonferroni test was used for all statistical analyses.