Figure 1. A model for RNAi/PTGS pathways in vegetative cells in N. crassa.
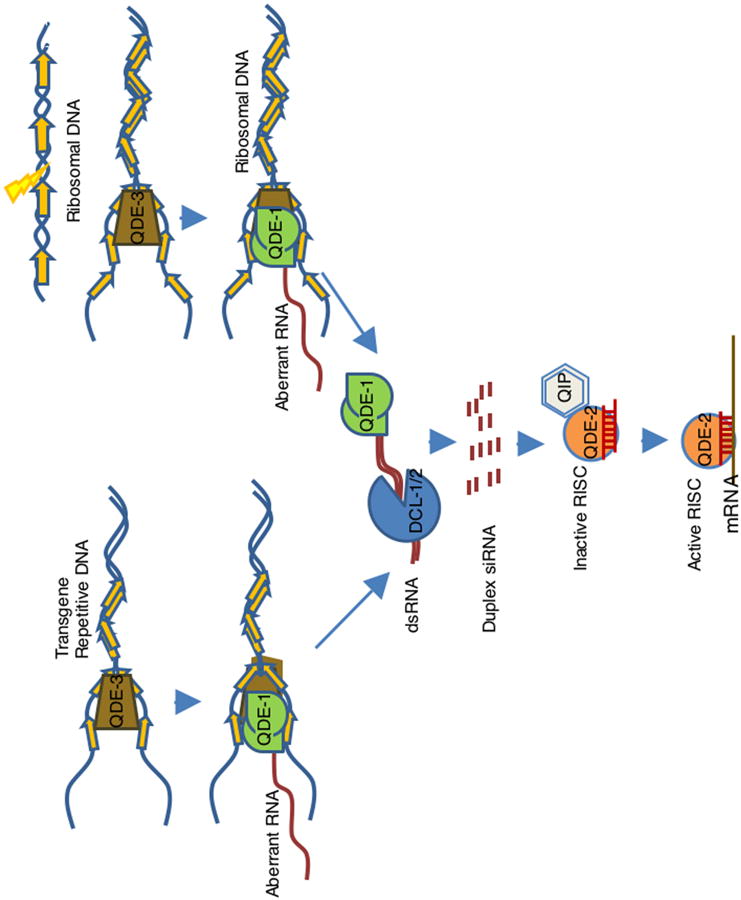
This model puts together (a) transgene-induced PTGS (quelling), (b) dsRNA-induced PTGS and (c) DNA-damage induced qiRNA pathway. Transgenes (quelling) or DNA-damage induce the synthesis of aberrant RNAs by the DdRP activity of QDE-1 facilitated by QDE-3 and RPA, where QDE-3 could probably resolve into ssDNA the complex DNA structures at the transgenic locus created upon tandem integration or caused by DNA damage, and RPA could recruit QDE-1 to the ssDNA. The aberrant RNA is then transcribed into dsRNAs by the RdRP activity of QDE-1. The Dicer proteins DCL-1 and DCL-2 cleave the dsRNAs, formed by QDE-1 or expressed directly from dsRNA-expressing constructs, into approximately 25 nt siRNA duplexes (or 20-21nt qiRNAs for DNA damage induced dsRNA), which are then loaded onto the RNA-induced silencing complex (RISC) containing QDE-2 and QIP. QDE-2 and QIPconvert the siRNA duplex into the mature siRNA, resulting in RISC activation to silence targets with homology to the siRNA. qiRNA bind to the core component of QDE-2, and play roles in response to DNA damage, but it is not known if qiRNAs function through the RISC containg QDE-2 and QIP.
