Figure 4.
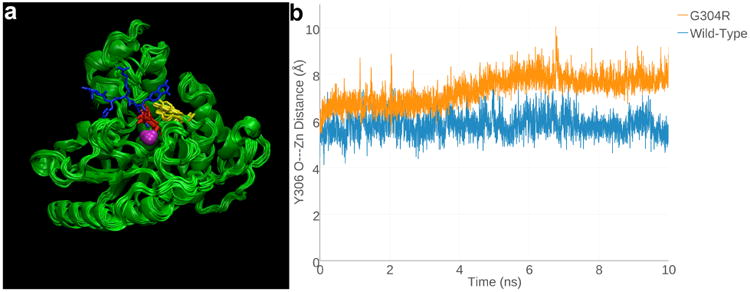
(a) Superimposed 1-ns snapshots from the 10-ns MD simulation of Zn2+-bound G304R HDAC8, with a tetrapeptide assay substrate (blue) superimposed for reference (from the structure of the H143 HDAC8-substrate complex, PDB accession code 3EWF). Zn2+ is a magenta sphere, R304 is red, and Y306 is yellow. (b) Y306 fluctuates ∼2Å away from the “in” conformation required for catalysis in G304R HDAC8 relative to its fluctuations in the wild-type enzyme over the course of the 10-ns MD simulation.
