Figure 6.
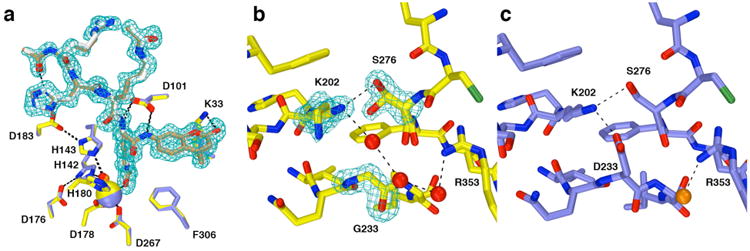
(a) Comparison of substrate binding in the D233G-Y306F HDAC8–substrate complex (C = yellow (protein) or tan (substrate), N = dark blue, O = red, Zn2+= yellow sphere, water = red sphere, monomer B) and the Y306F HDAC8–substrate complex (C = blue (protein) or gray (substrate), N = dark blue, O = red, Zn2+= blue sphere, water = orange sphere, monomer A, PDB accession code 2V5W). Metal coordination and hydrogen bond interactions are shown as solid black and dashed lines, respectively. The simulated annealing omit map (contoured at 3.0σ) shows a nearly fully ordered tetrapeptide substrate bound in the active site of D233G-Y306F HDAC8. (b) Simulated annealing omit maps of the D233G-Y306F HDAC8–substrate complex (monomer B, color coded as in (a)) showing the mutated residue G233 (contoured at 5.0σ) and the side chains of K202 and S276 (contoured at 3.0σ), each of which adopt two conformations. An ordered water molecule fills the void created by the D233G mutation and hydrogen bonds with K202 and a second water molecule. (c) Structure of the Y306F HDAC8–substrate complex (monomer A, color coded as in (a), PDB accession code 2V5W). Comparison with (b) illustrates structural changes resulting from the D233G mutation.
