Figure 7.
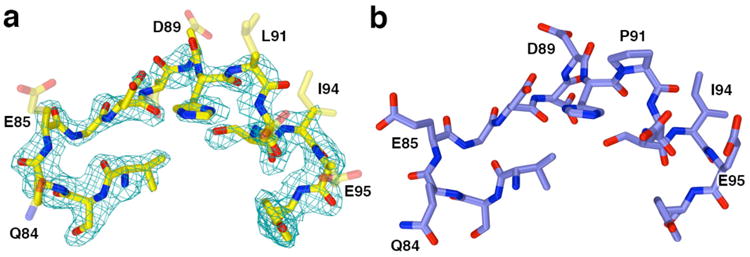
(a) Simulated annealing omit map of the L2 loop segment flanking residue 91 in the P91L-Y306F HDAC8–substrate complex (monomer A, contoured at 2.7σ) indicates that the side chains of residues Q84, E85, D88, D89, L91, D92, I94, and E95 are partially or completely disordered (these side chains are represented as transparent sticks). For comparison, the structure of the Y306F HDAC8–substrate complex (monomer A, PDB accession code 2V5W) is shown in (b). Color codes are as follows: C = yellow (P91L-Y306F HDAC8) or blue (Y306F HDAC8), N = dark blue, O = red.
