Figure 5.
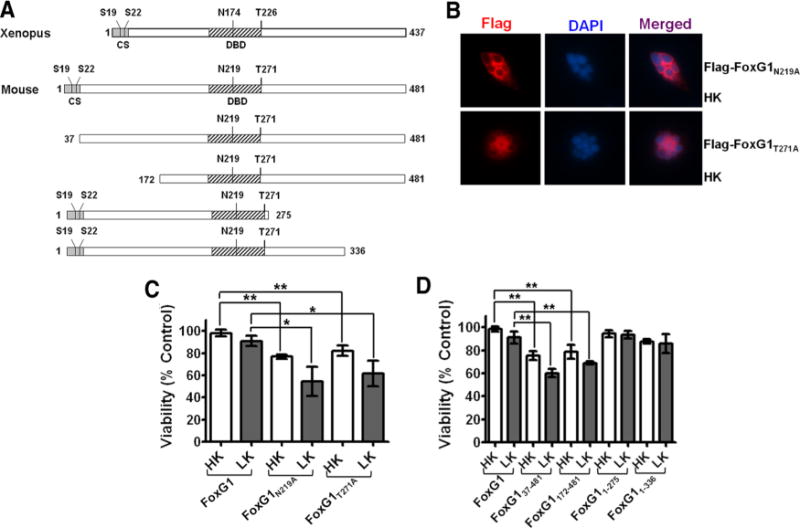
Sites or domains responsible for FoxG1 mediated neuronal survival. A, Schematic diagram of the wild-type FoxG1 protein from Xenopus [which was used by Regad et al. (2007)] and the mouse FoxG1 protein used in this study. Position of relevant amino acids as well as the conserved sequence region (CS) and the DBD is indicated. Also shown are the various FoxG1 deletion constructs used in this study. B, Immunocytochemistry result of CGN in HK transfected with Flag-FoxG1N219A and Flag-FoxG1T271A. C, CGNs were transfected with plasmids expressing wild-type FoxG1-Flag, the Akt phosphorylation site mutant Flag-FoxG1T271A, and Flag-FoxG1N219A, a mutant construct in which an aspartate residue essential for DNA binding is substituted with alanine. The neurons were then switched to HK or LK medium for 24 h, after which viability of transfected neurons was quantified. Both Flag-FoxG1N219A and Flag-FoxG1T271A failed to protect neurons from LK-induced death. Results are from three different experiments performed in duplicate. **p< 0.01 as compared to FoxG1-transfected CGNs in HK. *p< 0.05 as compared to FoxG1-transfected neurons in LK. D, Quantification of neuronal survival with the various FoxG1-Flag deletion constructs. Deletion of N-terminus regions destroys the ability of FoxG1 to prevent neuronal death in LK. Moreover, these constructs inhibit survival in HK, possibly through a dominant-negative mechanism. In contrast, C-terminus deletion constructs had no effect on HK-mediated survival. Results come from three different experiments performed in duplicate. **p< 0.01 as compared to FoxG1-transfected CGNs in HK or LK as indicated in the graph by bars.
