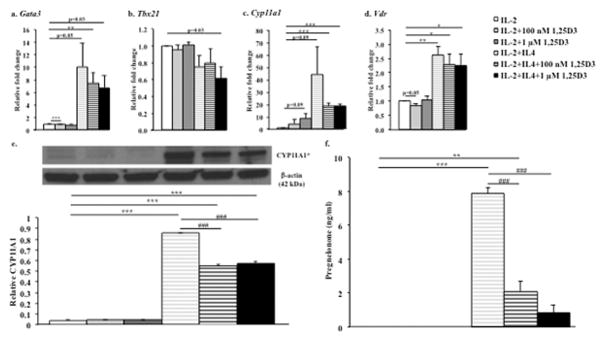
Figure 2. 1,25D3 treatment of CD8+ T cells alters gene expression of transcription factors and the functional activity of CYP11A1.
Gene expression of (a) Gata3, (b) Tbx21, (c) Cyp11a1, and (d) Vdr was measured by quantitative PCR (qPCR) in CD8+ T cells in IL-2 or IL-2+IL-4 in the presence of absence of 100 nM or 1 μM 1,25D3. Results (relative fold change+SEM) are from three independent experiments. (e) CYP11A1 protein levels (mean+SEM) detected by immunoblot analyses and densitometry of autoradiographs in CD8+ T cells differentiated in IL-2 or IL-2+IL-4 in the presence of absence of 100 nM or 1 μM 1,25D3. Results are from three independent experiments, *calculated molecular weight (MW) for CYP11A1 (13363-1-AP, Proteintech, Chicago, IL): 60 kDa, observed MW: 49 kDa, (f) Pregnenolone levels (mean+SEM) determined by ELISA in supernatants from CD8+ T cells differentiated in IL-2 or IL-2+IL-4 in the presence or absence of 100 nM or 1 μM 1,25D3. Results are from six independent experiments. Linear mixed models were employed; pairwise comparisons were performed using t-tests derived from these models. *p<0.05, **p<0.01, ***p<0.001 compared to the IL-2 group, ###p<0.001 compared to the IL-2+IL-4 group, these p-values remained significant after correction for multiple comparisons (Benjamini-Hochberg57 correction); p-values that did not reach the threshold p-value after adjustment for multiple comparisons are shown numerically.