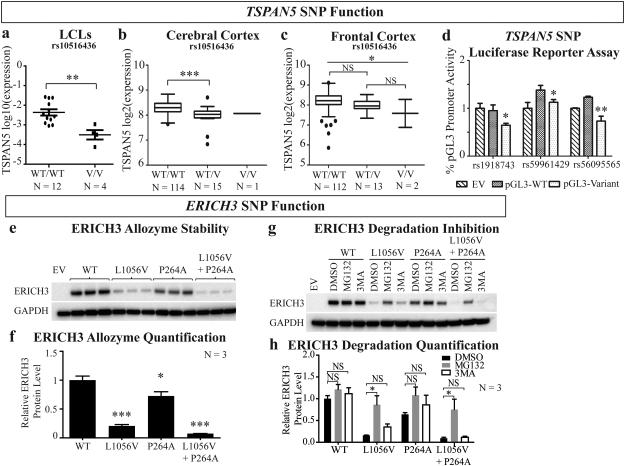
Figure 3. TSPAN5 and ERICH3 SNP function.
TSPAN5 expression is decreased for cells of tissues homozygous for variant (V/V) or with heterozygous (WT/V) SNP genotypes as compared to homozygous wild type (WT/WT) in (a) LCLs, (b) cerebral cortex and (c) frontal cortex. (d) Luciferase assay results comparing WT and variant SNP genotypes (rs1918743, rs59961429, and rs56095565) effects on transcriptional activities indicate decreased transcription for the variant SNPs in SK-N-BE(2) neuroblastoma cells; (e) ERICH3 plasmids that were WT or contained one or both of the non-synonymous SNPs (rs11580409 and rs11210490) were expressed in HEK-293T/17 cells. Both P264A (rs11210490) and L1056V (rs11580409) were associated with decreased protein levels as compared with WT, but L1056V was associated with a much greater decrease in protein level. (f) Quantification of ERICH3 protein relative to the GAPDH control for the ERICH3 Western blots shown in (e). (g) Plasmids encoding ERICH3 allozymes that were WT or contained one or both of the amino acid substitutions (P264A and L1056V) were expressed in HEK-293T/17 cells with and without a protease inhibitor (MG132) or an autophagy inhibitor (3MA). MG132 prevented ERICH3 SNP-dependent protein degradation but 3MA did not. (h) Quantification of proteasome and autophagy inhibition of the ERICH3 allozyme degradation studies shown in (g). EV = Empty Vector; NS = non-significant; * = p < 0.05; ** = p < 0.01; *** = p<0.0001.