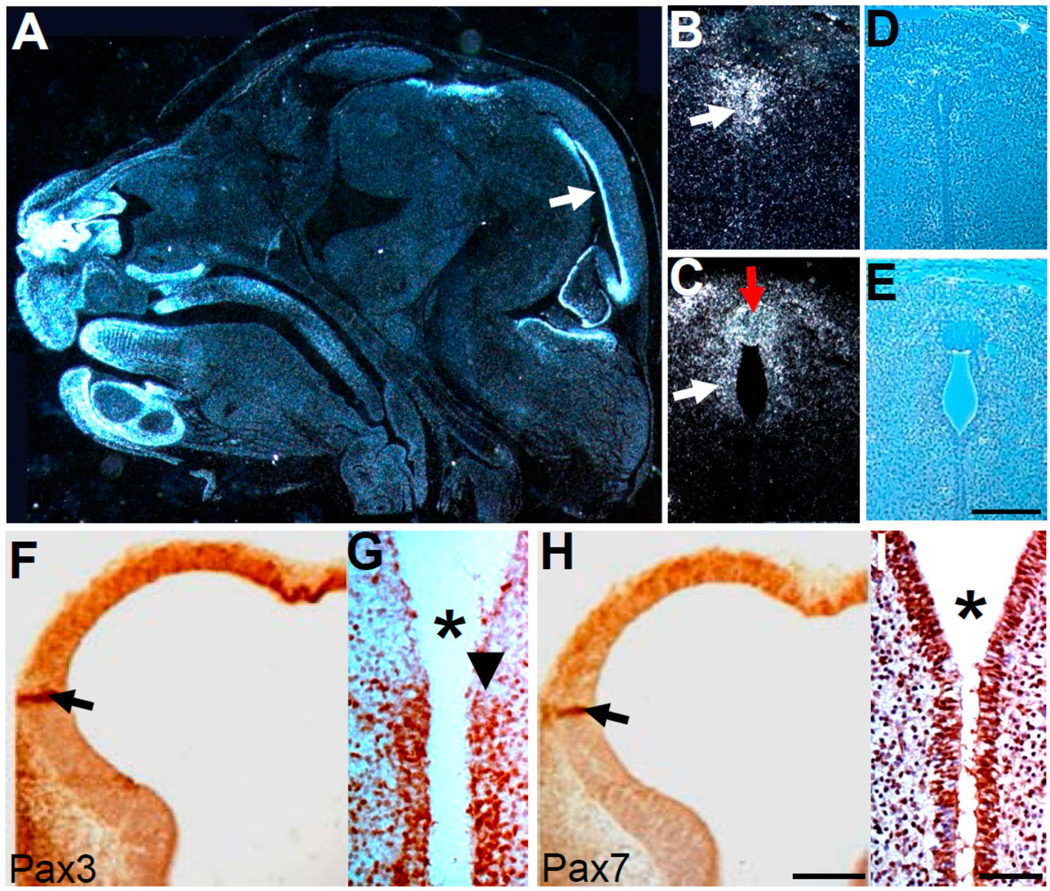
Figure 7.
Analysis of Pax3 and Pax7 spatiotemporal expression patterns, Cre expression limits and cell proliferation during cerebral ventricular development. (A–C) In situ hybridization detection of Pax3 mRNA in E15.5 wild-type heads show active expression in ventricular epithelium (arrow) and multiple craniofacial structures (A). Coronal E14.5 sections cut at the subcommissural organ (SCO, indicated by the red arrow in C) level of Pax3flox/flox/Wnt1-Cre mutant (B) and control (C) reveal Pax3 is still expressed in the ventricular epithelium (white arrow) comparable to control (C), despite collapse of the ventricular opening in mutants; (D,E) Phase contrast images of B,C, illustrating the lack of a distinct subcommissural organ in the mutant D); (F,H) Immunohistochemical detection of Pax3 (F) and Pax7 (H) protein on adjacent telencephalic vesicle sections at E10, reveals co-localization in the majority of the neuroepithelium, with especially robust expression (brown) in the lamina terminalis. The black arrows point to intense artificial staining due to tissue folding; (G,I) Immunohistochemical detection beta-galactosidase expression following lineage mapping at E13.5 for Pax7−Cre/R26r (G) and Wnt1-Cre/R26r (I) following Cre-mediated activation of R26r reporter. While Wnt1-Cre marks the ependymal layer from third ventricle (3v) through aqueduct (I), Pax7−Cre only marks the ependymal layer of 3v (arrow points to the 3v-aqueduct junction and asterisk indicates aqueduct). Scale bars: B–E = 100 µm; F,H = 200 µm; G,I = 50 µm.