Abstract
Skeletal muscle mitochondria are arranged as a reticulum. Insight into the functional characteristics of such structure is achieved by viewing the network as consisting of "subsarcolemmal" (SS) and "intermyofibrillar" (IMF) regions. During the decades, most, but not all, published studies have reported higher (sometimes over 2-fold) enzyme and enzyme-pathway protein-specific activities in IMF compared to SS mitochondria. We tested the hypothesis that non-mitochondrial protein contamination might account for much of the apparently lower specific activities of isolated SS mitochondria. Mouse gastrocnemii (n = 6) were suspended in isolation medium, minced, and homogenized according to procedures typically used to isolate SS mitochondria. However, the supernatant fraction, collected after the first slow-speed (800×g) centrifugation, was divided equally: one sample was exposed to nagarse (MITO+), while the other was not (MITO−). Nagarse treatment reduced total protein yield by 25%, while it increased protein-specific respiration rates (nmol O2 min−1 mg−1), by 38% under "resting" (state 4) and by 84% under maximal (state 3) conditions. Nagarse therefore increased the respiratory control ratio (state 3/state 4) by 30%. In addition, the ADP/O ratio was increased by 9% and the activity of citrate synthase (U/mg) was 49% higher. Mass spectrometry analysis indicated that the MITO+ preparation contained less contamination from non-mitochondrial proteins. We conclude that nagarse treatment of SS mitochondria removes not only non-mitochondrial proteins but also the protein of damaged mitochondria, improves indices of functional integrity, and the resulting protein-specific activities.
Keywords: nagarse, oxygen consumption, skeletal muscle, mass spectrometry, mitochondrial respiration
Introduction
Mitochondrial oxidative phosphorylation fulfills two critical metabolic objectives: 1) synthesizing ATP at the rate demanded by cellular ATP-utilizing processes and 2) maintaining a robust cellular energy status (low cytosolic [ADP] and high ATP/ADP ratio). Almost 40 years ago Skulachev's laboratory provided evidence that mitochondria in mammalian striated muscle are structured as a continuous network or reticulum [1]. Recently, Glancy et al. [2] provided compelling corroborative evidence for this model. Further, their report supports one of the fundamental hypotheses advanced by Skulachev over 40 years ago [1], that the reticular arrangement provides the ability to rapidly transfer intracellular energy by propagating the protonmotive force (Δp) from one region of the network to another [1, 3, 4]. According to this concept, the fraction of the network located near the plasma membrane ("subsarcolemmal" or SS mitochondria) contains protein stoichiometry particularly suited for Δp development, while the region deeper within the myocyte, the intermyofibrillar (IMF) fraction, is particularly tailored for transducing Δp into ATP synthesis and export. Reticular structure therefore facilitates meeting the demands for both rapid ATP turnover and the defense of cellular energetic status. These older and more recent findings bracket decades of research supporting the mitochondrial reticulum concept.
About the time Skulachev's paper demonstrated the mitochondrial reticulum in rat diaphragm [1], Palmer et al. described procedures for independently isolating SS and IMF mitochondrial populations from rat heart [5]. Briefly, the tissue was minced, suspended in buffer, mechanically disrupted (liberating SS mitochondria) and centrifuged at slow speed, leaving SS mitochondria in the supernatant, while the IMF mitochondria were pelleted with the myofibrils. This first supernatant was then used to isolate SS mitochondria with high-speed spins. In the parallel IMF isolation procedure, the myofibrillar pellet was resuspended and incubated with the proteolytic enzyme nagarse to digest myofibrillar proteins and liberate the IMF mitochondria. Next, a second slow spin would yield a supernatant containing IMF mitochondria. The IMF supernatant was then centrifuged at high speed to pellet and wash the IMF mitochondria. These careful and detailed studies by Palmer et al. [5, 6] provided convincing electron microscopic evidence that SS and IMF fractions were independently isolated, and their procedures became the generally accepted methodology upon which subsequent work was based. However, a curious pattern reported by Palmer et al., and many studies that followed [5–10], was that essentially all activities of individual enzymes and/or oxidative enzyme pathways were uniformly higher in the IMF fraction compared to the SS, in some cases over 2-fold higher. This consistent finding raises obvious questions: If SS and IMF mitochondria are simply part of a continuous network, then how could all protein-specific activities be higher in IMF? Another, related, question is: Where and by what mechanism would the network transition from this SS (lower) to IMF (higher) protein-specific activity? One obvious alternative explanation is that these apparent differences simply reflect experimental artifact. SS mitochondria are isolated in the absence of nagarse exposure, while IMF isolation fundamentally depends on the nagarse incubation. Because nagarse treatment is the glaring difference between the two procedures, we tested the simple hypothesis that non-mitochondrial protein contamination accounts for the apparently lower specific activities of isolated SS mitochondria. Our data support this hypothesis and moreover advance the concept that nagarse treatment may also remove the protein within and perhaps also attached to damaged mitochondria that, left undigested, would otherwise diminish indices of mitochondrial structural and functional integrity.
Material and methods
Animal and muscle preparation
All procedures were in accordance with the guidelines regarding the care and use of animals by the Institutional Animal Care and Use Committee at Mayo Clinic. A total of six C57BL/6J mice on standard chow diet and water ad libitum were used for all experiments. Mice ranged in age from 8 to 12 weeks. On the day of the experiment, the mice were euthanized by isoflourane inhalation and cervical dislocation. The left and right gastrocnemii were removed and immediately placed on an ice-cold petri dish, which was pre-rinsed with ice-cold modified Chappell-Perry Medium I (Solution I; mM): 100 KCl, 40 Tris-HCl, 10 Tris-Base, 5 MgCl2, 1 EDTA, 1 ATP, pH 7.5. After removing the blood, fat and visible connective tissues, the gastrocnemii were placed into a pre-massed beaker containing 2 ml of ice-cold Solution I, reweighed, and the wet muscle mass was calculated. The mean gastrocnemii mass was 138.7±10.5 mg.
Isolation of mitochondria
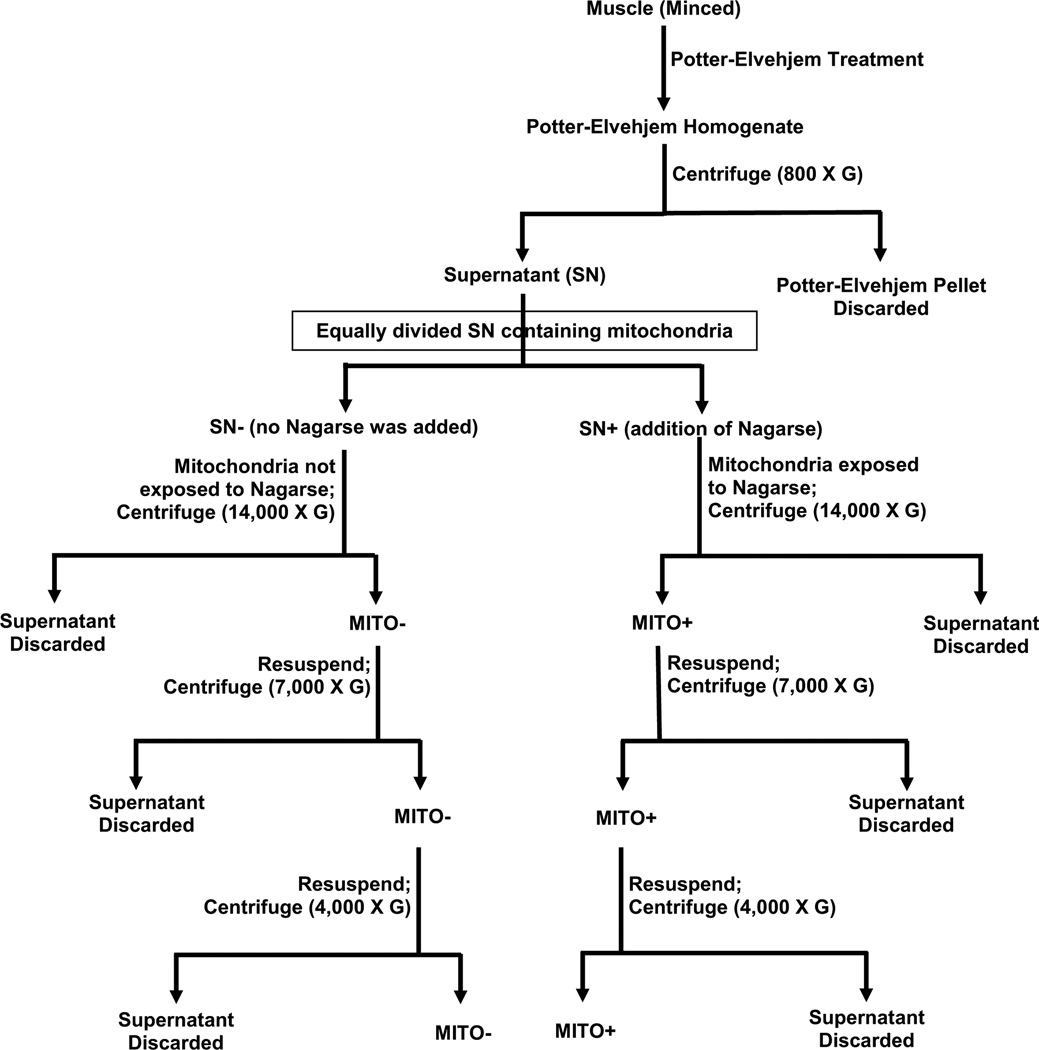
Mitochondria were prepared according to the outline in Fig. 1; all procedures were carried out on ice or at 4°C and all centrifugations were 10 min in duration. Muscles were cleaned, weighed, minced with scissors in 9 volumes of ice-cold Solution I, and gently homogenized by hand using a ground glass-to-glass Potter-Elvehjem homogenizer (5 passes). The homogenate was centrifuged at 800 × g to obtain the supernatant (SN) containing mechanically released (SS) mitochondria. This SN was divided equally into two 1.5 ml homogenization tubes. Nagarse (bacterial proteinase Type XXIV, Sigma, P-8038) was prepared using Solution I (5 mg.g−1 ww) and added to one tube (SN+). An equal volume of Solution I containing no nagarse, was added to the other tube (SN-). After 7 min incubation, 1 ml of Solution I was added to each tube (this terminated nagarse digestion in the SN+ tube). The SN+ and SN- samples were then centrifuged at 14,000 × g to obtain two mitochondrial pellets, one exposed to nagarse (MITO+) and the other not exposed (MITO−). After discarding the supernatant, each of the mitochondrial pellets were re-suspended in 0.5 ml of Solution II (mM): 100 KCl, 40 Tris-HCl, 10 Tris-Base, 5 MgCl2, 1 EDTA, 0.2 ATP, and 1.5% BSA, pH 7.5. Following centrifugation at 7000×g, the supernatants were discarded and the mitochondrial pellets were re-suspended in 0.5 ml of Solution III (identical to Solution II, but without BSA). After the final centrifugation at 4000 × g and removal of supernatant, the two final mitochondrial pellets were each re-suspended in identical volumes of mannitol-sucrose buffer containing (mM) 220 Mannitol, 70 Sucrose, 10 Tris-HCl, 1 EGTA, pH 7.40. The volume of buffer used to resuspend each mitochondrial preparation was 1 ul per mg of original wet muscle. The protein content in the final mitochondrial preparations was determined by the method of Lowry [11]. All protein concentrations reported below and in the tables relate exclusively to the final mitochondrial preparations.
Figure 1. Isolation and treatment of the muscle mitochondria.
A flow diagram depicting the preparation of mechanically released mitochondrial sample that was subsequently either exposed or not to the enzyme nagarse. Details are given in the “Methods” (SN, supernatant).
Citrate synthase assay
Citrate synthase (CS) activity was determined spectrophotometrically at 37°C by the method of Srere [12], as previously described [13]. In these assays aliquots of mitochondrial suspensions, stored at −80°C, were assayed in buffer that included 0.05% Triton detergent to disperse the mitochondrial inner membrane and eliminate all enzyme latency.
Assays for mitochondrial O2 consumption and ATP production
Freshly isolated mitochondria were assayed for O2 consumption rate (Jo) and ATP production rate (Jp). O2 consumption was measured polarographically in a respiration chamber (Hansatech Instruments, Norfolk, UK) at 37°C following general procedures we have previously described [13]. Mitochondrial respiration was fueled with the substrate combination pyruvate (1 mM) + malate (1 mM) + glutamate (10 mM) (PMG). Aliquots, typically 20 ul in volume, of mitochondrial suspension were added to 250 µl of respiration medium adapted from Wanders et al. [14], which contained (in mM) 100 KCl, 50 MOPS, 10 K2PO4, 10 MgCl2, 1 EGTA, and 0.2% BSA, pH 7.00 [13]. Next, the PMG substrate combination was added and State 2 Jo was followed (respiration due primarily to proton leak). The addition of ADP to give a final concentration of 0.67 mM stimulated state 3, (maximal) Jo. Phosphorylation of this ADP resulted in state 4 Jo [15], and the respiratory control ratio (RCR) was calculated as state 3 Jo/state 4 Jo. The ADP/O ratio was determined as previously described [15]. The State 3 (maximal) rate of ATP production was calculated as the product of state 3 Jo times the ADP/O (taking the 2:1 molecular to atomic oxygen stoichiometry into account).
Protein separation by SDS-PAGE
Final MITO+ and MITO− suspensions were diluted 1:1 in 2× Laemmli sample buffer before running on a SDS-PAGE gel. Laemmli sample buffer containing β-mercaptoethanol was prepared according to manufacturer’s instructions (Bio-Rad, Hercules, CA). Samples were heated at 95°C for 5 minutes and then loaded onto a pre-cast 10% SDS-PAGE gel (Bio-Rad, Hercules, CA). The gel was run at 60 volts for 30 minutes, 110 volts for 60 minutes and then 150 volts for 10 minutes. Performing gel electrophoresis under these conditions allowed for adequate separation, visualization, and the ability to compare the MITO+ versus MITO− samples. Proteins were visualized using Coomassie blue. Gel image was captured using an ImageQuant LAS 4000 (GE Healthcare Life Sciences).
Protein identification and quantification by mass spectrometry
To obtain an insight into the abundance of mitochondrial versus non-mitochondrial proteins contained in each of the MITO+ and MITO− preparations, aliquots from the samples analyzed on SDS-PAGE were also analyzed by mass spectrometry to identify and quantify proteins contained in each mitochondrial preparation. The mass spectrometry procedure was also used to identify and quantify the protein nagarse in the same preparations.
In-solution Digest
A volume of isolated mitochondrial preparation, either treated or not treated with the enzyme nagarse, was added to a 9 times volume of dilution buffer [DB - 10% acetonitrile (ACN) and 25 mM Tris-HCl pH 8.5]. 8 µl trypsin (Sigma; St. Louis, MO) at 0.200 ug/ml was added to the protein sample and was allowed to incubate for 16 hours at 37°C with gentle shaking, followed by addition of 50 µl 5% formic acid (FA) to halt the digestion. The resulting peptides were prepared for sample analysis similar to a previously published protocol [16]. In brief, a stop-and-go extraction tip (StageTip) [17], was fitted with two C18 disk plugs using a customized tipping syringe [17]. The Stage Tip was activated with methanol, washed in 100 µl buffer B (0.1% FA, 80% ACN), and equilibrated in 100 ul buffer A (0.1% FA) twice. The peptides were then loaded onto the activated Stage Tip, washed twice in 100 ul buffer A, followed by elution in 50 ul Buffer B. The eluate was dried by vacuum centrifugation and stored at −80°C prior to use. 6 µl of 0.1% FA (v/v) was added to re-suspend the dried samples, followed by sonication for 2 min. The sonicated samples were briefly centrifuged and 1 µl of sample was subsequently analyzed by mass spectrometry as described below.
Mass spectrometry
HPLC-ESI-MS/MSn was performed on a Thermo Electron Orbitrap Elite Velos Pro fitted with an EASY source (Thermo Electron, San Jose, CA). NanoLC was performed using a DIONEX/Thermo NCS-3500RS UltiMate 3000 with an EASY Spray column (Thermo Electron, 50cm × 75-um inner diameter, packed with PepMap RSLC C18 material, 2 um); loading phase for 15 min; mobile phase, linear gradient of 1–37% ACN in 0.1% FA in 150 min, followed by a step to 95% ACN in 0.1% FA over 5 min, hold 10 min, and then a step to 1% ACN in 0.1% FA over 1 min and a final hold for 19 min (total run 200 min); Buffer A = 0.1% FA in 100% H2O; Buffer B = 0.1% FA in 100% ACN; flow rate, 300 nl/min. All solvents were mass spectrometry grade. A “top 15” data-dependent MS/MS analysis was performed (acquisition of a full scan spectrum followed by collision-induced dissociation mass spectra of the 15 most abundant ions in the survey scan).
Database search
Tandem mass spectra were extracted by ProteoWizard msConvert, version 3 [18] using the default settings. Charge state deconvolution and deisotoping were not performed. All MS/MS spectra were analyzed using Mascot (Matrix Science, London, UK; version 2.4.1). Mascot was set up to search the SwissProt_02_2015 database (16706 entries, mus musculus) assuming the digestion enzyme trypsin and a maximum of 2 missed cleavages permitted. Mascot searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 10.0 PPM. Phosphorylation of serine, threonine, and tyrosine as well as oxidation of methionine was specified in Mascot as a variable modification while no fixed modifications were indicated.
Criteria for protein identification
Scaffold (version Scaffold_4.3.4, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability. Peptide Probabilities were assigned by the Peptide Prophet algorithm [19] with Scaffold delta-mass correction as well as by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [20]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Quantification of mitochondrial and non-mitochondrial proteins
Progenesis software (Nonlinear Dynamics; Progenesis QI; Quayside, Newcastle Upon Tyne, UK) was used to quantify abundance of peptides associated with each protein. Quantification using non-conflicting peptides was selected to generate the protein report for the peptide ion abundance associated with each identified protein. Accession numbers were imported into SwissProt_02_2015 database and used to assign the subcellular location of the identified proteins (i.e. mitochondrial, non-mitochondrial). Peptide ion abundances for all mitochondrial and all non-mitochondrial proteins in each of the MITO+ and MITO− samples were added to calculate total mitochondrial and total non-mitochondrial protein abundances, respectively.
Quantification of the nagarse abundance
To quantify the abundance of nagarse present in the mitochondrial preparations treated/not treated with nagarse, we used the same approach described in the “Database search,” section, but with the following exception: Mascot was set up to search the SwissProt_02_2015 database for bacillus licheniformis. We used the unique and stable identifier from the SwissProt_02_2015 database for nagarse, Q65LP7, to perform all queries. We also used the corresponding mnemonic identifier of the UniprotKB entry, Q65LP7_BACLD to perform all protein queries. The ion abundance for the peptides associated with the enzyme nagarse within each of the mitochondrial preparations were compared with the total ion abundance for all the proteins within each of the mitochondrial preparations treated/not treated with nagarse from the Progenesis protein reports. The total ion abundances are in reference to alignments in Mascot search using SwissProt_02_2015 database for mus musculus. The ion abundances of the peptides associated with the enzyme nagarse are in reference to alignments in Mascot search using SwissProt_02_2015 database for bacillus licheniformis.
Statistical analyses
Differences between MITO+ and MITO− across variables of interest were compared using paired t-test. Data are reported as means ± SEM. Significance was set at P <0.05.
Results
Protein content of final mitochondrial suspension
Isolating mitochondria with nagarse decreased the protein concentration of the final mitochondrial suspension by 30%. The MITO+ preparation was 2.12 ± 0.23 mg protein.ml−1 compared to 2.80 ± 0.37 mg.ml−1 in MITO−. When these protein concentrations (mg/ml) are multiplied by their respective suspension volumes (ml), the total protein yield (mg “mitochondrial” protein) of the isolation procedure is calculated. When this is done, the MITO+ protein yield was 0.290 ± 0.03 mg, compared to a MITO− value of 0.386 ± 0.05 mg (Table 1). Thus, nagarse exposure reduced, by 25%, the total protein isolated (Table 1).
Table 1.
Total protein yield, enzyme specific activities, and indices of functional integrity of mitochondria isolated without and with nagarse
| MITO− | MITO+ | MITO+/MITO− (Fold Δ) |
P Value | |
|---|---|---|---|---|
| Total protein yield mg | 0.386 ± 0.05 | 0.290 ± 0.03 | 0.75 | 0.02 |
| Citrate synthase activity nmol·min−1·mg−1 | 827.4 ± 65.4 | 1232.9 ± 83.5 | 1.49 | 0.0001 |
| State 3 JO nmol·min−1·mg−1 | 77.6 ± 8.6 | 142.6 ± 16.8 | 1.84 | 0.003 |
| State 4 JO nmol·min−1·mg−1 | 22.5 ± 2.4 | 31.0 ± 3.5 | 1.38 | 0.01 |
| State 3 JP nmol·min−1·mg−1 | 384.9 ± 36.9 | 771.4 ± 83.4 | 2.00 | 0.002 |
| RCR | 3.6 ± 0.4 | 4.6 ± 0.4 | 1.30 | 0.002 |
| ADP/O | 2.5 ± 0.1 | 2.7 ± 0.1 | 1.09 | 0.02 |
Values are means ± SE; MITO+, mitochondria samples treated with nagarse; MITO−, mitochondria samples not treated with nagarse; total protein yield was calculated as the product of the protein concentration in the final mitochondrial suspension times the suspension volume; state 3 Jo, state 3 O2 consumption rate; state 4 Jo, state 4 O2 consumption rate; state 3 Jp, state 3 ATP production rate. RCR, respiratory control ratio, is State 3 (maximum) Jo divided by State 4 (resting) Jo; State 3 Jp is the product of State 3 Jo and the ADP/O ratio.
Citrate synthase activity
Citrate synthase, the first enzyme of the citric acid cycle, is a sturdy matrix enzyme often used to assess muscle mitochondrial content. Expressed per mg protein in the final mitochondrial suspension, citrate synthase activity (U.mg−1) was 49% higher when nagarse was included in the isolation procedure (Table 1). However, the total citrate synthase activity recovered in the final mitochondrial suspension, calculated by multiplying the U.mg−1 by the total protein (mg) yield in the final suspension (see above), was not different in MITO+ (366.7 ± 53.0 nmol.min−1) compared to MITO− (329.3 ± 54.1 nmol.min−1) (Table 2).
Table 2.
Total yield of citrate synthase and respiratory activity of mitochondria isolated without and with nagarse
| MITO− | MITO+ | MITO+/MITO− (Fold Δ) |
P Value | |
|---|---|---|---|---|
| Citrate synthase activity nmol·min−1 | 329.3 ± 54.1 | 366.7 ± 53.0 | 1.11 | 0.11 |
| State 3 JO nmol·min−1 | 31.1 ± 5.7 | 42.5 ± 7.5 | 1.37 | 0.01 |
| State 4 JO nmol·min−1 | 9.1 ± 1.8 | 9.1 ± 1.3 | 1.00 | 0.98 |
| State 3 JP nmol·min−1 | 153.5 ± 27.5 | 229.5 ± 38.8 | 1.49 | 0.01 |
Values are means ± SE; MITO+, mitochondria samples treated with nagarse; MITO−, mitochondria samples not treated with nagarse; total yield of activity was calculated as the product of specific activity times the total protein yield (see mean values in Table 1); state 3 Jo, state 3 O2 consumption rate; state 4 Jo, state 4 O2 consumption rate; state 3 Jp, state 3 ATP production rate.
Respiratory rates and respiratory control ratio
Mitochondrial O2 consumption (Jo) due to the combustion of Pyruvate + Malate + Glutamate (PMG Jo) requires the entire oxidative pathway of fully intact mitochondria, including several soluble matrix cofactors such as Coenzyme A and NAD+. Saturating ADP was added to stimulate the maximum O2 consumption rate (state 3 Jo). When mitochondria phosphorylate all of the added ADP to ATP, they transition to “resting” (state 4) respiration.
Table 1 reports that nagarse treatment increased both state 3 Jo (by 84%) and state 4 Jo (by 38%), when these values are expressed per mg protein (nmol O2.min−1.mg−1). Thus, nagarse treatment especially increased State 3 Jo, while it increased State 4 more modestly. As a result, nagarse increased the respiratory control ratio (RCR = State 3 Jo/State 4 Jo) by 30% (Table 1). The total yield of State 3 Jo activity recovered in the final mitochondrial suspension was calculated as above for citrate synthase, by multiplying the respiration rates per mg by the total protein (mg) yield in the final suspension. Unlike citrate synthase, which was not statistically different, total state 3 Jo was 37% higher in MITO+ (42.5 ± 7.5 nmol.min−1) compared to MITO− (31.1 ± 5.7 nmol.min−1) (Table 2). The total yield of State 4 Jo activity was essentially identical in MITO− and MITO+ (Table 2).
ADP/O coupling and maximum ATP production
Nagarse exposure modestly improved, by 9%, the ADP/O ratio, 2.7 ± 0.1 vs. 2.5 ± 0.1, in MITO+ vs. MITO−, respectively (Table 1). The product of State 3 Jo times the ADP/O yields the State 3 ATP production rate (State 3 Jp). Because both factors were elevated by nagarse treatment, State 3 Jp was dramatically (100%) higher in MITO+ compared to MITO−, 771.4 ± 83.4 vs. 384.9 ± 36.9 nmol ATP.min−1.mg−1 respectively (Table 1). Again, multiplying these protein-specific State 3 Jp values by the total protein (mg) yield indicated that nagarse exposure increased the total yield of State 3 Jp by 49%: 229.5 ± 38.8 nmol ATP/min in MITO+ vs. 153.5 ± 27.5 nmol ATP/min in MITO− (Table 2).
Protein detection by SDS-PAGE and identification and quantification by mass spectrometry
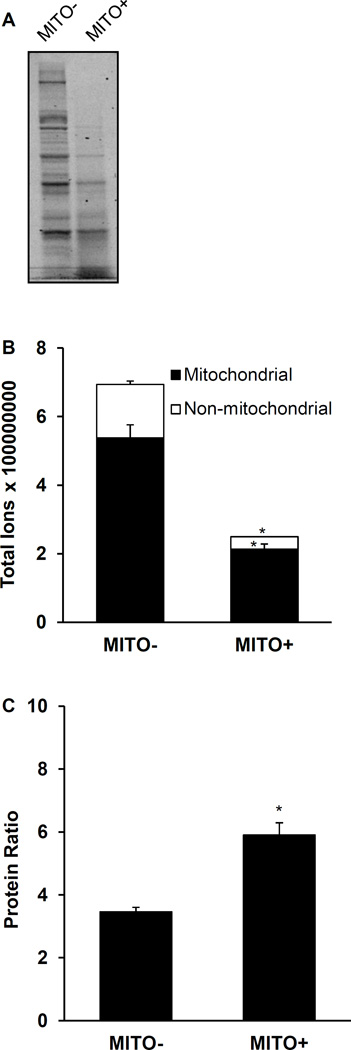
Consistent with the Lowry protein determinations, nagarse treatment decreased the visually apparent protein detected using SDS-PAGE in representative MITO+ vs. MITO− preparations (Fig. 2A). Mass spectrometry analysis of all samples also showed less protein, including mitochondrial protein, content in the MITO+ suspensions (Fig. 2B). In MITO+ preparations approximately 83% of the total protein content was identified as mitochondrial proteins. In contrast, in MITO− preparations mitochondrial proteins could account for only approximately 73% of total protein. The ratio of identified MITO proteins versus non-MITO proteins was higher in MITO+ samples compared to MITO− (Fig. 2C).
Figure 2. Protein abundance in mitochondrial suspensions isolated without and with nagarse.
Protein abundance detected by SDS-PAGE/Coomassie Blue (A), HPLC-ESI-MS/MS quantitative proteomics analysis to determine peptide ion abundance of all mitochondrial and all non-mitochondrial proteins in MITO+ and MITO− samples (B), and protein ratio of mitochondrial-to-non-mitochondrial protein in the same samples (C). Values are means ± SE; MITO+, mitochondria samples treated with nagarse; MITO−, mitochondria samples not treated with nagarse; *P < 0.05 between MITO+ and MITO−.
Nagarse (apr; subtilisin Carlsberg; EC:3.4.21.62; UniProtKB accession number, Q65LP7) was essentially absent in the final suspensions of mitochondria subjected to nagarse treatment. In MITO+ preparations the tryptic peptide abundance of nagarse represented only 0.008% of the total peptide abundance. In MITO− this value was 0.0002% (data not shown).
Discussion
The major findings reported here are that nagarse exposure substantially reduced (by 25%) the protein yield in the final mitochondrial suspension, and proteomic analysis indicated that the elimination of nonmitochondrial protein accounted for much of this reduction. By removing non-mitochondrial protein, nagarse treatment increased mitochondrial enzyme and enzyme pathway protein-specific activities. Expressed per mg protein, nagarse treatment increased citrate synthase activity by 49% and State 3 Jo even more dramatically, by 84%. Moreover, nagarse also modestly increased the ADP/O ratio, so that, per mg protein, State 3 Jp improved by 100%. The data generally indicate that nagarse treatment effectively removes contaminating non-mitochondrial proteins and substantially improves indices of mitochondrial catalytic potential and energetic coupling.
The total isolated CS activity was not significantly affected by nagarse treatment, while nagarse increased the total yield of State 3 Jo by 37%, and State 3 Jp by 49%. The spectrophotometric assay of citrate synthase activity versus the polarographic assay of mitochondrial oxidative phosphorylation fueled by PMG evaluate two vastly different parameters of mitochondrial structure and function. Citrate synthase is a high activity matrix enzyme with well-known stability [21], and is routinely measured in either whole tissue homogenates or mitochondrial suspensions in the presence of a detergent such as Triton X-100, as it was in the present study. In marked contrast, the oxidation of the substrate combination PMG, whether the mitochondria are “resting” or stimulated by saturating ADP, requires, at the least, the entire integrated oxidative pathway and, in particular, robust inner membrane structural integrity. It can be argued that the different outcomes of the CS and Jo assays suggest that nagarse treatment somehow modified the first, slow speed, supernatant in a way that protected mitochondrial units (vesicles) from damage during the subsequent high speed centrifugation and resuspension steps of the isolation procedure. This interpretation is developed below.
Whether mitochondria are partially damaged, have been briefly opened and then resealed, or are fully intact, citrate synthase activity may well remain unaffected. Thus, it can be envisioned that in the absence of nagarse treatment some mitochondria, both fully intact and partially damaged, in the first supernatant were subsequently further damaged during the course of the isolation procedure, but they nevertheless carried citrate synthase activity into the final suspension. Nagarse treatment, on the other hand, would be expected to proteolytically remove the activity of exposed citrate synthase in damaged mitochondria. The data of Table 2 indicate that these intact mitochondria in the final MITO+ suspension carry roughly similar citrate synthase as the MITO−. The net effect is that the total yield of citrate synthase activity in the final suspension is not significantly different. However, in MITO+ more of this citrate synthase activity resides in fully intact mitochondria, which are capable of coupled oxidative phosphorylation.
The total yield of State 3 Jo was 37% higher and State 3 Jp activity was 49% higher in MITO+ final suspensions. In contrast, the total yield of State 4 Jo was essentially identical in the two preparations. Nagarse in the first supernatant may have somehow protected against mitochondrial damage during subsequent centrifugation and resuspension steps, perhaps by digesting non-mitochondrial proteins in some type of linkage [22, 23] to mitochondria. In the abstance of nagarse treatment, during the high-speed centrifugations and pellet resuspensions these linkages somehow increased the likelihood of structural insult to mitochondrial vesicles. The damage was not severe enough to liberate citrate synthase from its matrix binding [21], but was sufficient to preclude the development of the very high driving forces required for oxidative phosphorylation. Damaged organelles in MITO− preparations are consistent with the lower RCR and ADP/O values observed in MITO−. It therefore appears that nagarse promoted the release of mitochondria from non-mitochondrial protein, which was digested and resulted in less protein yield and less damage to mitochondria during isolation.
Mass spectrometry data further support this interpretation. MITO− preparations had much greater abundance of non-mitochondrial proteins. In nagarse treated preparations, both mitochondrial and nonmitochondrial protein yields were reduced (Fig. 2B). This suggests that sufficiently damaged mitochondria in the first supernatant were exposed to nagarse entry/attack, digested and did not make it into the final suspension.
The mitochondria within a muscle cell are interconnected as a reticulum (network) [1–3, 24, 25]. Mechanical homogenization, however gentle, of muscle tissue therefore must destroy this reticular structure, because electron micrographs of isolated mitochondria, for example those of Palmer et al. [5], clearly show the isolated organelles as individual vesicles. During homogenization, as these vesicles form, some membrane damage and leakage of essential matrix cofactors (NAD, CoA, adenylates, etc.) into the isolation medium would be expected [26]. Nevertheless, when these procedures are performed carefully by experienced hands, and particularly when nagarse is included in the isolation procedure, the resulting vesicles are capable of nearly matching the maximum O2 consumption rates [27, 28] and ATP free energy [29–31] measured in intact muscle using Fick O2 mass balance and 31P-MRS, respectively. Moreover, the isolated mitochondria also control respiration and ATP production over the same range of energy phosphate levels observed in vivo [13, 30, 32, 33]. Thus, mitochondria isolated using mechanical homogenization and exposure to protease can be nearly as functional as mitochondria in vivo.
Conclusions
Nagarse treatment of mitochondria removes non-mitochondrial proteins as well as proteins of damaged mitochondria, and improves indices of functional integrity and resulting protein-specific activities.
Highlights.
Exposure of mitochondria to nagarse removes non-mitochondrial proteins
Exposure of mitochondria to nagarse removes protein of damaged mitochondria
Treating mitochondria with nagarse improves indices of functional integrity
Acknowledgments
The authors thank James Krantz and Alicia Polito at Mayo Clinic in Arizona for skillful technical assistance in conducting the mice experiments. The study was supported by a NIH/NIDDK grant DK094062 to CSK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bakeeva LE, Chentsov Yu S, Skulachev VP. Mitochondrial framework (reticulum mitochondriale) in rat diaphragm muscle. Biochim Biophys Acta. 1978;501:349–369. doi: 10.1016/0005-2728(78)90104-4. [DOI] [PubMed] [Google Scholar]
- 2.Glancy B, Hartnell LM, Malide D, Yu ZX, Combs CA, Connelly PS, Subramaniam S, Balaban RS. Mitochondrial reticulum for cellular energy distribution in muscle. Nature. 2015;523:617–620. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amchenkova AA, Bakeeva LE, Chentsov YS, Skulachev VP, Zorov DB. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J. Cell Biol. 1988;107:481–495. doi: 10.1083/jcb.107.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skulachev VP. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 2001;26:23–29. doi: 10.1016/s0968-0004(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 5.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- 6.Palmer JW, Tandler B, Hoppel CL. Biochemical differences between subsarcolemmal and interfibrillar mitochondria from rat cardiac muscle: effects of procedural manipulations. Arch. Biochem. Biophys. 1985;236:691–702. doi: 10.1016/0003-9861(85)90675-7. [DOI] [PubMed] [Google Scholar]
- 7.Bizeau ME, Willis WT, Hazel JR. Differential responses to endurance training in subsarcolemmal and intermyofibrillar mitochondria. J Appl Physiol (1985) 1998;85:1279–1284. doi: 10.1152/jappl.1998.85.4.1279. [DOI] [PubMed] [Google Scholar]
- 8.Cogswell AM, Stevens RJ, Hood DA. Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. Am J Physiol. 1993;264:C383–C389. doi: 10.1152/ajpcell.1993.264.2.C383. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi M, Hood DA. Protein import into subsarcolemmal and intermyofibrillar skeletal muscle mitochondria. Differential import regulation in distinct subcellular regions. J Biol Chem. 1996;271:27285–27291. doi: 10.1074/jbc.271.44.27285. [DOI] [PubMed] [Google Scholar]
- 10.Schwerzmann K, Hoppeler H, Kayar SR, Weibel ER. Oxidative capacity of muscle and mitochondria: correlation of physiological, biochemical, and morphometric characteristics. Proc Natl Acad Sci U S A. 1989;86:1583–1587. doi: 10.1073/pnas.86.5.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 12.Srere PA. Citrate synthase. [EC 4.1.3.7. Citrate oxaloacetate-lyase (CoA-acetylating)] Methods in Enzymology C. 1969;13:3–11. [Google Scholar]
- 13.Lefort N, Glancy B, Bowen B, Willis WT, Bailowitz Z, De Filippis EA, Brophy C, Meyer C, Hojlund K, Yi Z, Mandarino LJ. Increased reactive oxygen species production and lower abundance of complex I subunits and carnitine palmitoyltransferase 1B protein despite normal mitochondrial respiration in insulin-resistant human skeletal muscle. Diabetes. 2010;59:2444–2452. doi: 10.2337/db10-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanders RJ, Groen AK, Van Roermund CW, Tager JM. Factors determining the relative contribution of the adenine-nucleotide translocator and the ADP-regenerating system to the control of oxidative phosphorylation in isolated rat-liver mitochondria. Eur J Biochem. 1984;142:417–424. doi: 10.1111/j.1432-1033.1984.tb08303.x. [DOI] [PubMed] [Google Scholar]
- 15.Estabrook R, Pullman M. Methods in Enzymology: Oxidation and Phosphorylation. New York: Academic Press; 1967. [Google Scholar]
- 16.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 17.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 18.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 21.D'Souza SF, Srere PA. Binding of citrate synthase to mitochondrial inner membranes. J Biol Chem. 1983;258:4706–4709. [PubMed] [Google Scholar]
- 22.Boncompagni S, Rossi AE, Micaroni M, Beznoussenko GV, Polishchuk RS, Dirksen RT, Protasi F. Mitochondria are linked to calcium stores in striated muscle by developmentally regulated tethering structures. Mol. Biol. Cell. 2009;20:1058–1067. doi: 10.1091/mbc.E08-07-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 24.Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacin M, Vidal H, Rivera F, Brand M, Zorzano A. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem. 2003;278:17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- 25.Kirkwood SP, Munn EA, Brooks GA. Mitochondrial reticulum in limb skeletal muscle. Am J Physiol. 1986;251:C395–C402. doi: 10.1152/ajpcell.1986.251.3.C395. [DOI] [PubMed] [Google Scholar]
- 26.Bremer J, Wojtczak A, Skrede S. The leakage and destruction of CoA in isolated mitochondria. Eur J Biochem. 1972;25:190–197. doi: 10.1111/j.1432-1033.1972.tb01684.x. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen UF, Rasmussen HN. Human skeletal muscle mitochondrial capacity. Acta Physiol Scand. 2000;168:473–480. doi: 10.1046/j.1365-201x.2000.00699.x. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen UF, Vielwerth SE, Rasmussen HN. Skeletal muscle bioenergetics: a comparative study of mitochondria isolated from pigeon pectoralis, rat soleus, rat biceps brachii, pig biceps femoris and human quadriceps. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2004;137:435–446. doi: 10.1016/j.cbpb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Hitchins S, Cieslar JM, Dobson GP. 31P NMR quantitation of phosphorus metabolites in rat heart and skeletal muscle in vivo. Am J Physiol Heart Circ Physiol. 2001;281:H882–H887. doi: 10.1152/ajpheart.2001.281.2.H882. [DOI] [PubMed] [Google Scholar]
- 30.Jeneson JA, Westerhoff HV, Brown TR, Van Echteld CJ, Berger R. Quasi-linear relationship between Gibbs free energy of ATP hydrolysis and power output in human forearm muscle. Am J Physiol. 1995;268:C1474–C1484. doi: 10.1152/ajpcell.1995.268.6.C1474. [DOI] [PubMed] [Google Scholar]
- 31.Kemp GJ, Meyerspeer M, Moser E. Absolute quantification of phosphorus metabolite concentrations in human muscle in vivo by 31P MRS: a quantitative review. NMR Biomed. 2007;20:555–565. doi: 10.1002/nbm.1192. [DOI] [PubMed] [Google Scholar]
- 32.Glancy B, Willis WT, Chess DJ, Balaban RS. Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry (Mosc) 2013;52:2793–2809. doi: 10.1021/bi3015983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messer JI, Jackman MR, Willis WT. Pyruvate and citric acid cycle carbon requirements in isolated skeletal muscle mitochondria. Am J Physiol Cell Physiol. 2004;286:C565–C572. doi: 10.1152/ajpcell.00146.2003. [DOI] [PubMed] [Google Scholar]