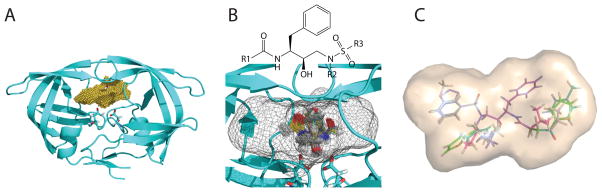
Figure 1.
Illustration of our computational approach for inhibitor design. (A) Grid generation: the binding pocket of each target structure (cyan cartoon) is partitioned into grids (in yellow) on which van der waals energies and continuum electrostatic potentials are calculated. (B) Scaffold placing: conformational choices of a scaffold are first determined by assessing discretized translational, rotational, and conformational rotamers on grids (the envelope of the grids is now in a mesh representation). (C) Functional group placing: combinations of functional-group rotamers are pruned efficiently for each scaffold conformation retained in Step B. Subsequently, the energetically most favorable ensemble of structures for each target structure is reranked with more accurate energy models hierarchically.