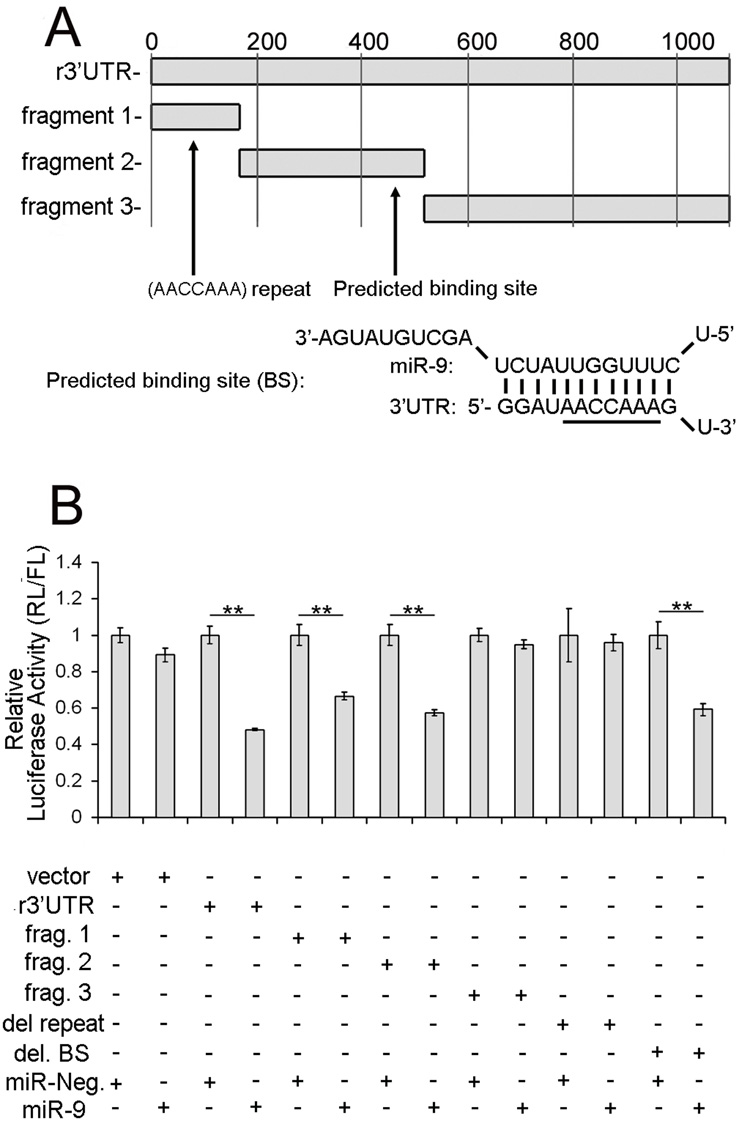
Figure 4. miR-9 interacts in vitro with the 3’UTR of PMP22.
(A) Luciferase constructs containing fragments of the 3’UTR. Non-overlapping segments of the 3’UTR are used to delineate the binding sites of miR-9. The r3’UTR construct contains the full length 3’UTR. The remaining constructs fragment 1 (nts 1–157), fragment 2 (nts 158–498) and fragment 3 (nts 499–1127) are segments of the full length 3’UTR. The predicted binding site (BS) is located in Fragment 2 and the predicted base pairing is shown. The (AACCAAA) sequence is underlined. Two additional (AACCAAA) sequences are found in Fragment 1.
(B) Activity of miR-9 on luciferase constructs. The plasmids are co-transfected with either miR-9 or miR-Neg, a scramble miRNA (negative control). miR-9 has no effect on the empty vector or the plasmid containing fragment 3 (Frag. 3). However, miR-9 does significantly reduce luciferase activity of constructs containing either the full length 3’UTR of PMP22 (r3’UTR), fragment 1 (Frag. 1) or fragment 2 (Frag. 2) (**p<0.01, Student’s t-test, as compared to Neg). miR-9 also down-regulates luciferase activity of PMP del. BS (corresponding to the 3’UTR of PMP22 with a deletion of the predicted binding site (BS)) (**p<0.01, Student’s t-test, as compared to Neg) while no difference is observed with the PMP del. repeat plasmid (3’UTR with a deletion of the (AACCAAA) repeat). Error bars represent the s.d. n=6.