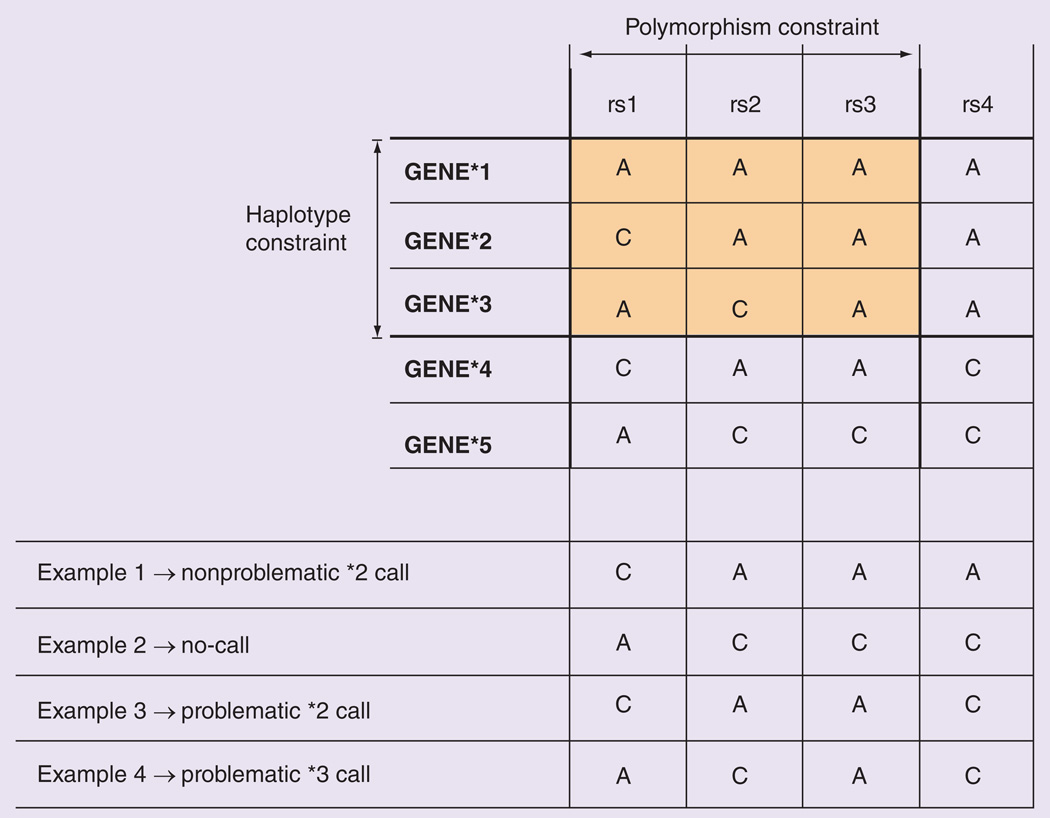
Figure 1. Illustration of the concept of ‘constrained views’ based on restricted sets of polymorphisms and haplotypes (top); examples of haplotype calls produced by the constrained view (bottom).
The constrained view only takes the highlighted variants into account for haplotype calling, excluding other polymorphisms and haplotypes in the full haplotype definition table. In this example, the assay tests three (rs1, rs2 and rs3) of the four variant sites and reports three (GENE*1, GENE*2 and GENE*3) of the five defined haplotypes. Therefore, while the hypothetical dataset contains results for all four variant sites (examples 1–4 at bottom), only three of them are used in haplotype assignment. In example 1, the assigned haplotype (GENE*2) is correct. A haplotype cannot be assigned for example 2 because the genotype patterns at rs1, rs2 and rs3 do not match any of the reported haplotype alleles (*1–*3). The assigned haplotypes for examples 3 and 4 are incorrect due to the assay’s constrained view regarding variant sites and/or defined haplotypes, and they are therefore categorized as ‘problematic calls’. Problematic calls are those in which a constrained view leads to calling a haplotype that is not called with the FULL view.