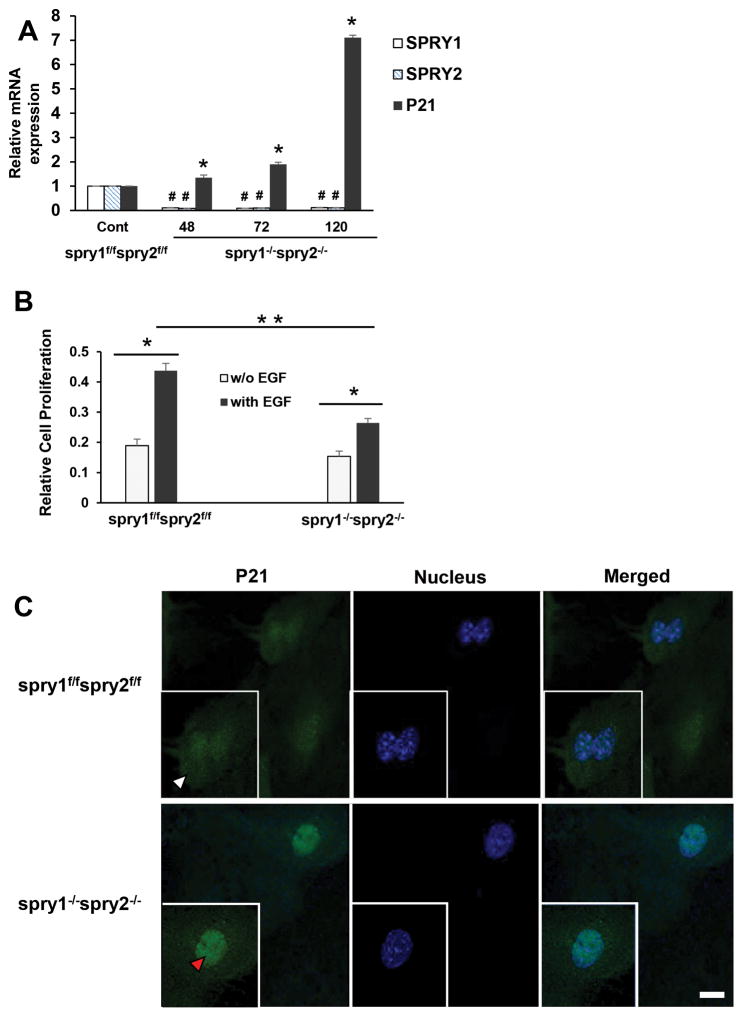
Fig. 8. Spry1 and Spry2 deletion in mouse embryonic fibroblasts upregulates nuclear p21 expression and inhibits EGF-dependent proliferation.
(A) Tamoxifen dependent recombination and a complete deletion of Spry1 and Spry2 in MEFs increases p21 expression. Spry1f/f;Spry2f/f and Spry1−/−;Spry2−/− MEFs were cultured for 48–120 hr. Total RNA was extracted and mRNA transcripts of SPRY1, SPRY2 and p21 were assessed by RTPCR. Data represents means ± SD of three independent experiments, #,*p< 0.05, compared to control Spry1f/f;Spry2f/f MEFs (B) Deletion of Spry1 and Spry2 in MEFs reduces EGF-dependent cell proliferation. Spry1f/f;Spry2f/f and Spry1−/−;Spry2−/− MEFs were plated and treated with vehicle or EGF (10ng/ml) for 48hr. Cell proliferation was assessed by MTT assay. Groups compared; vehicle and EGF treated Spry1f/f;Spry2f/f cells, vehicle and EGF treated Spry1−/−;Spry2−/− cells and EGF treated Spry1−/−;Spry2−/− cells compared to EGF treated Spry1f/f;Spry2f/f cells. Data represents means ± SD of three independent experiments, *p< 0.05 (one way ANOVA). (C) Representative confocal images showing the expression of p21 in Spry1f/f;Spry2f/f and Spry1−/−;Spry2−/− MEFs. Cells were fixed with cold methanol, permeabilized, and blocked with normal goat serum. Sequentially, the cells were incubated with mouse anti-p21 and Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin. The cells were then stained with DAPI to label the nuclei. Images were obtained using a Leica TCS SP8 confocal laser scanning microscope. Cytoplasmic (white arrow head) and nuclear (red arrow head) localization of p21 is shown. All images were collected using identical microscope settings and magnification. The panels are representative of at least three independent experiments in which similar patterns were obtained for the indicated cells.