Abstract
The anti-metabolite 5-fluorouracil (5-FU) is employed clinically to manage solid tumors including colorectal and breast cancer. Intracellular metabolites of 5-FU can exert cytotoxic effects via inhibition of thymidylate synthetase, or through incorporation into RNA and DNA, events that ultimately activate apoptosis. In this review, we cover the current data implicating DNA repair processes in cellular responsiveness to 5-FU treatment. Evidence points to roles for base excision repair (BER) and mismatch repair (MMR). However, mechanistic details remain unexplained, and other pathways have not been exhaustively interrogated. Homologous recombination is of particular interest, because it resolves unrepaired DNA intermediates not properly dealt with by BER or MMR. Furthermore, crosstalk among DNA repair pathways and S-phase checkpoint signaling has not been examined. Ongoing efforts aim to design approaches and reagents that (i) approximate repair capacity and (ii) mediate strategic regulation of DNA repair in order to improve the efficacy of current anti-cancer treatments.
Keywords: colorectal cancer, chemotherapy, DNA damage, base excision repair, mismatch repair, homologous recombination
1. Introduction
Based on the initial observations of Rutman et al. [1] and Heidelberger et al. [2] that rat hepatoma cells utilize uracil to a greater extent in nucleic acid biosynthesis than non-malignant cells, it became of great medical interest to identify uracil analogs that exhibited “selective” anti-cancer activity. Shortly thereafter, the synthesis of 5-fluorouracil (5-FU; Figure 1) [3] and its efficacy as a potential anti-tumor drug was reported [4]. This compound, as well as the nucleoside analog 5-fluoro-2′-deoxyuridine (FdUrd; Figure 1), are part of a class of cytotoxic drugs known as anti-metabolites, which have been integrated into numerous clinical trials and found to exhibit anti-tumor activity in patients. Today, 5-FU is widely used in the treatment of solid tumors, including of the breast, gastrointestinal system (colon, rectum, anus, esophagus, pancreas and stomach), head and neck, and ovary [5]. Most notably, 5-FU is routinely employed in the management of colorectal cancer via one of two FDA-approved first line combinatorial chemotherapy regimes, abbreviated FOLFOX and FOLFIRI, which involve intravenous administration of the fluorinated base analog (Figure 1).
Figure 1. Structure of 5-FU (left) and FdUrd (right).
The fluorine atom at the 5 position of the pyrimidine ring distinguishes 5-FU from uracil (hydrogen at the 5 position) and thymine (methyl group at the 5 position).
Despite the recent exciting advances in targeted therapeutics, such as the development of kinase inhibitors and monoclonal antibodies that specifically block the growth of cancer cells (e.g. imatinib mesylate (Gleevec™) or trastuzumab (Herceptin™)), traditional cytotoxics including 5-FU continue to be used in combination chemotherapy, primarily as a means of combating drug-resistant malignant cell populations to which all treatment regimes old and new fall prey [6,7]. The pharmacokinetic profiles and side effects of traditional cytotoxics are also well understood through decades of use. As aspects of cancer management focus on patient quality of life and move towards outpatient treatment paradigms, orally administered anti-cancer drugs are a priority. It is noteworthy in this regard that capecitabine (Xeloda™), which is commonly used to treat breast and colorectal cancer, is an orally available pro-drug of 5-FU [8].
There are multiple pathways involved in the activation and degradation of 5-FU (see Section 2). Depending on the metabolic path, 5-FU and its metabolites can exert anti-proliferative effects through inhibition of thymidylate synthetase (TS) and/or incorporation into RNA and DNA [9]. It remains debated as to the relative contribution of each of these cellular targets to the anti-tumor activity and side effects seen in patients. This review will focus on the DNA repair processes associated with responding to 5-FU in chromosomal DNA, a topic that has received comparatively little attention until recently.
2. 5-FU Metabolism and its Directed Effects
Because 5-FU is a structural analog of uracil and thymine, many of the enzymes that participate in uracil or thymine metabolism also effectively metabolize 5-FU, topics that have been extensively studied and previously reviewed [10]. This brief summary highlights pathways to DNA incorporation.
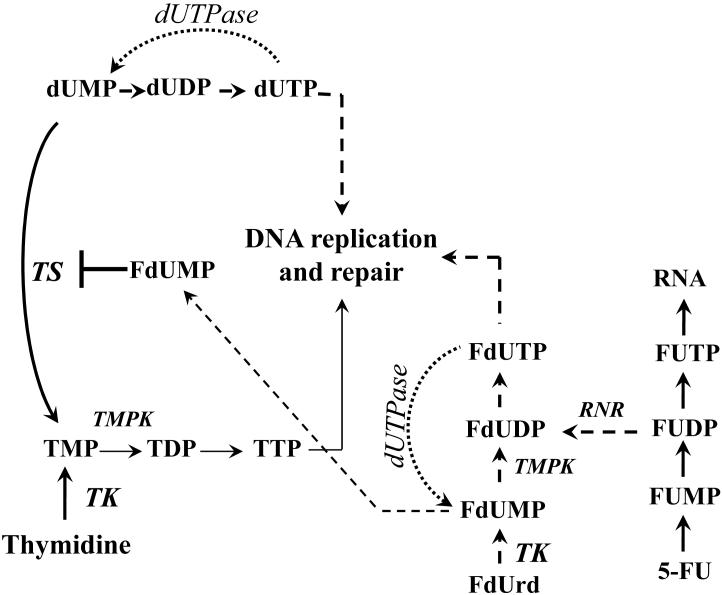
5-FU and FdUrd rapidly enter cells by a facilitated transmembrane carrier system. Once in the cell, conversion to nucleotides promotes intracellular retention and further metabolism (Figure 2). Uridine phosphorylase and orotate phosphoribosyltransferase convert 5-FU into the ribonucleoside or ribonucleotide, respectively. Thymidine phosphorylase can also salvage 5-FU into the deoxynucleoside, FdUrd. Kinases convert FUrd to FUMP and to FUDP, which provides a branch point in metabolite fate. FUDP is phosphorylated to FUTP, which is a substrate for RNA polymerases. FUDP can also be converted to the deoxynucleotide (FdUDP) by ribonucleotide reductase (RNR). FdUDP is further phosphorylated into FdUTP, which is a substrate for DNA polymerases (discussed in Section 2.1).
Figure 2. Simplified scheme of 5-FU and FdUrd metabolism.
Enzymes are italicized. TS, thymidylate synthase provides the only de novo source of TMP. dUTPase, dUTP nucleotidohydrolase prevents dUTP and FdUTP accumulation. TK, thymidine kinase salvages thymidine and FdUrd. FdUMP suicide inhibits TS. TMPK, thymidylate kinase, phosphorylates TMP and FdUMP. RNR, ribonucleotide reductase, converts FUDP and UDP to FdUDP and dUDP, respectively.
The enzyme dUTP pyrophosphatase (dUTPase) performs an absolutely essential function to prevent genomic uracil incorporation by catalyzing the hydrolysis of dUTP into dUMP (Figure 2); E. coli and S. cerevisiae lacking dUTPase are inviable [11,12]. dUTPase also breaks down FdUTP to FdUMP, which is a noteworthy aspect of 5-FU metabolism. While breaking down FdUTP prevents 5-FU from being incorporated into DNA, it creates FdUMP in the process. FdUMP forms an irreversible ternary complex with TS (a classic biochemistry textbook example of suicide inhibition), the enzyme that converts dUMP to TMP using N5,N10-methylenetetrahydrofolate as a coenzyme to establish thymidylate nucleotides essential for DNA replication [13]. When FdUrd is used in cell culture experiments, thymidine kinase (TK) efficiently converts FdUrd into FdUMP, driving the generally accepted conclusion that FdUrd primarily exerts its toxicity via TS-directed effects. It is presumed that thymidylate kinase is capable of phosphorylating FdUMP to form FdUDP, which speculatively could counteract the TS inhibitory consequences of FdUMP.
Suicide inhibition of TS by FdUMP also causes a drop in TTP, which has several effects that can influence 5-FU metabolism. First, dUMP accumulates, resulting in a higher intracellular concentration of dUTP and FdUTP, which can overwhelm dUTPase and become available for incorporation into the genome by DNA polymerases. Second, TTP feedback inhibits TK and allosterically regulates RNR (TTP normally increases dGDP formation and decreases dUDP formation). Loss of this feedback inhibition presumably increases the conversion of FdUrd to FdUMP by TK and the conversion of FUDP to FdUDP by RNR. Thus, there are several interrelated events that can lead to the introduction of 5-FU into DNA (see Section 2.1). We refer readers to other reviews that cover aspects of 5-FU metabolism, including incorporation of 5-FU into RNA and suicide inhibition of TS [9,10,14]. This review will focus on more recent studies that have examined the repair machinery involved in the cellular responses to 5-FU once inserted into DNA.
2.1 5-FU in genomic DNA
Because 5-FU and uracil metabolism are so intertwined, genomic incorporation of both bases can result from 5-FU treatment (see Figure 2). In particular, dUTP is readily incorporated opposite adenine during DNA replication by a number of polymerases, which do not appear to discriminate between dUTP and TTP. Moreover, DNA polymerase α, the enzyme responsible for synthesis of a chimeric RNA-DNA primer for leading and lagging strand replication, and the DNA repair polymerase, POLβ (see Section 3.1 for additional details), reportedly incorporate FdUTP into DNA opposite adenine with an efficiency similar to dUTP and TTP [15,16]. To our knowledge, neither the processive replicative polymerases (POLδ/ε) nor any of the more specialized DNA polymerases discovered since 1997 have been evaluated for their efficiency to insert FdUTP during DNA synthesis, but undoubtedly, many of them are capable of such activity.
A number of groups have examined incorporation of radiolabeled 5-FU into DNA using 5-FU or FdUrd in a number of cell culture systems [17-26]. Several of the studies detected substantial amounts of genomic 5-FU, although it is not surprising that the amounts varied given the many experimental murine and human cell culture models examined and the multiple metabolic steps required. In some cases, genomic 5-FU was nearly undetectable if FdUrd was used [18,22]. Although earlier studies provide equivocal evidence that genomic 5-FU incorporation contributes to toxicity [17-26], it was pointed out that the rates of incorporation versus the excision efficiency were not determined [10]. Thus, the dynamic interplay between incorporation and DNA repair in dictating the steady state level of base damage was not examined. We discuss next the DNA repair pathways implicated in the recognition of genomic 5-FU.
3. Base Excision Repair and 5-FU Resistance
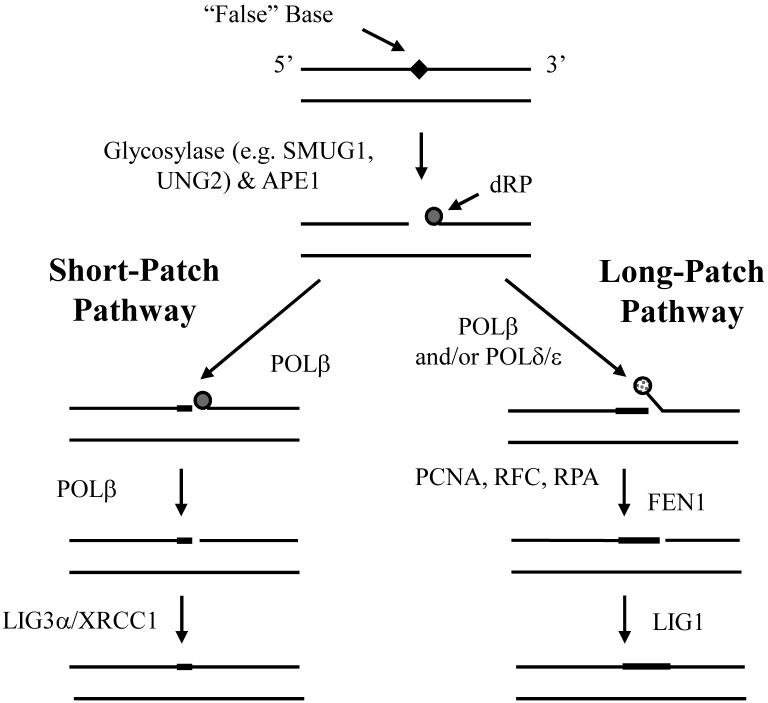
Base excision repair (BER) copes with specific forms of endogenous DNA damage. In particular, BER is the primary pathway for removing various types of oxidative, alkylative, and spontaneous hydrolytic DNA base and sugar products. The major steps of BER involve the following: (1) removal of a modified or inappropriate base, such as uracil, by a DNA glycosylase, (2) cleavage of the phosphodiester backbone at the resulting AP site by an endonuclease or lyase, (3) clean-up of the 3′ or 5′ terminal end, (4) replacement of the excised nucleotide by a polymerase, and (5) sealing of the final DNA nick by a ligase. The molecular events of BER and the predominant mammalian protein participants are depicted in Figure 3 [27,28].
Figure 3. The major enzymatic steps and proteins of mammalian BER.
BER is typically initiated by the removal of an inappropriate (e.g. uracil, 8-oxoguanine or certain mismatches) or “false” base, such as 5-fluorouracil, by a lesion specific DNA glycosylase. Following base excision, the resulting abasic site is most often incised by the major AP endonuclease, APE1, to create a strand break with a 5′-deoxyribose phosphate (dRP) residue. At this point, depending on the nature of the 5′-terminal end and other factors (reviewed in [27]), the DNA gap is restored via either short-patch (left) or long-patch (right) BER. In the former situation, the 5′-dRP residue is excised by the lyase activity of DNA POLb and the single nucleotide gap is filled by the same enzyme. Subsequently, a complex of XRCC1 and DNA ligase IIIα (LIG3) seals the remaining nick. In the case of long-patch BER, the 5′-terminal blocking fragment is ultimately removed by a flap endonuclease (FEN1) following strand-displacement synthesis by POLb and/or POLd/e. After excision of the flap DNA structure, the nick is sealed by DNA LIG1. PCNA, RFC and RPA help facilitate the long-patch repair response. See text for additional details.
There are four different known proteins in the human genome with uracil DNA glycosylase (UDG) activity. Note that the abbreviation UDG refers to biochemical activity, i.e. the ability to catalyze cleavage of the N-glycosidic bond, releasing the base from the sugar phosphodiester backbone (Figure 3), whereas abbreviations below refer to specific loci and polypeptides. Biochemical characterization of the four UDGs suggests specialized roles that combat two sources of uracil introduction into the genome, i.e. hydrolytic deamination of cytosine (giving rise to U:G pairs) and incorporation of dUMP during replication (generating U:A pairs), reviewed elsewhere in detail [29]. Briefly, the UNG genetic locus encodes mitochondrial (UNG1) and nuclear (UNG2) forms of UDGs, with UNG2 appearing to account for the bulk of cellular UDG activity [29]. The SMUG1 genetic locus encodes a glycosylase that has been proposed to serve as a backup for UNG in uracil excision, although SMUG1 also releases a broader range of damaged pyrimidine bases not excised by UNG [30]. TDG (thymine DNA glycosylase) and MBD4 (also known as MED1) appear to counteract cytosine or 5-methyl-cytosine deamination products in double stranded DNA, while TDG can also remove other types of damaged bases, most notably 3,N4 ethenocytosine [31].
3.1 5-FU excision by DNA glycosylases
The first evidence suggesting a role for BER in the cellular response to 5-FU was reported in 1980 [32]. In this study, both the bacterial and human (presumably UNG2) UDG was found to excise 5-FU, albeit with slightly less efficiency than uracil, from plasmid substrates harboring multiple tritium-labeled base lesions. This in vitro analysis was later confirmed using synthetic oligonucleotide duplexes that contained a single defined, site-specific 5-FU [33]; this study found that both purified E. coli and human UDG exhibited a 10 to 18-fold increase in KM for the 5-FU:A substrate relative to the U:A duplex, with little difference in Vmax. Since then, purified recombinant MED1 [34], TDG [35] and SMUG1 [30] have also been shown to remove 5-FU from synthetic DNA substrates in vitro. In the case of MBD4, this excision function is specific for 5-FU opposite guanine, a pairing con figuration for which there is no obvious mechanism of formation as will be discussed below (see Section 4.1). TDG is able to remove 5-FU opposite either guanine or adenine, as well as from single-stranded DNA, which stands in surprising contrast to the strict requirement of the enzyme for an opposing guanine when excising uracil or 3,N4 ethenocytosine [35]. SMUG1 excised 5-FU opposite adenine, but was not tested against other base partners [30]. This study also confirmed that UNG2 displayed a much stronger preference for uracil than 5-FU. Using human cell nuclear extracts and covalently closed circular DNA plasmids, 5-FU:G repair was found to be largely dependent on TDG and UNG2, whereas 5-FU:A pairs were processed mainly by UNG2 [36]. In these experiments, MBD4 and SMUG1 did not detectably contribute to 5-FU removal, yet given the in vitro excision activities of the recombinant proteins, could not be excluded from being involved in 5-FU metabolism in vivo.
In recent years, studies have begun to examine the biological involvement of specific mammalian BER components in the 5-FU response. In light of the biochemical studies described above, it was natural to suspect UNG. Yet surprisingly, several investigations have concluded using various approaches that UNG does not influence the cytotoxicity of 5-FU [37-39]. In particular, Ung +/+ and Ung -/- murine embryonic fibroblasts (MEFs) showed almost no difference in the lethal effects of 5-FU or FdUrd, despite an increased accumulation of uracil in Ung -/- cells [38]. In addition, expression of a protein inhibitor of UNG (i.e. Ugi) in HEK293 cells did not affect the toxicity of 5-FU or FdUrd despite a substantial increase in genomic uracil following treatment [39]. Thus, UNG activity or elevated uracil in DNA does not appear to contribute significantly to cellular sensitivity to 5-FU, although this does not rule out the possible involvement of other UDGs.
Indeed, work by Barnes and colleagues indicates that the SMUG1 glycosylase, not UNG, functions predominantly in cellular 5-FU repair, despite the fact that these enzymes possess comparable activities on 5-FU:A substrates in vitro [37]. In particular, the authors found that genomic 5-FU accumulates specifically in SMUG1-deficient MEFs, but not in Ung-/- MEFs, and that SMUG1-defective cells are uniquely sensitive to 5-FU treatment. One concerning aspect of the study was that the MEFs (regardless of genotype) displayed a greater sensitivity to 5-FU than FdUrd, whereas in nearly all studies using human cell lines FdUrd is at least 10-fold more toxic than 5-FU. That withstanding, the overall picture suggests that the excision activity of SMUG1, and not UNG, is protective against toxicity caused by genomic 5-FU. The results also imply that SMUG1 upregulation might serve as a means of developing tumor resistance to 5-FU treatment.
Cells deficient in the MBD4 (a.k.a. MED1) DNA glycosylase have been reported to be resistant to 5-FU [40,41]. This observation was extended to in vivo studies of Mbd4 -/- mice, in which it was found that apoptosis induced by 5-FU treatment in the small intestine was reduced in knockout animals relative to their wild-type counterparts [41]. This at first glance would appear to run contrary to the hypothesis that 5-FU in DNA is toxic. However, there is a connection between MBD4 and mismatch repair (MMR), a pathway known to promote cell death in response to DNA damage (see Section 4.1), worth emphasizing. Specifically, MBD4 has been shown to interact with MLH1, a key component of MMR [42]. In addition, defects in MMR are associated with hereditary non-polyposis colorectal cancer (HNPCC), i.e. cancers characterized by high microsatellite instability (MSI-H) [43], and mutations in the MBD4 gene have been reported in human colorectal cancers found to exhibit MSI-H [44-46]. While the links between MMR and genomic 5-FU will be discussed in more detail in Section 4, we note here that a deficiency in MED1 may result in an impaired MMR-dependent cell death response, leading to the observed increased resistance to 5-FU exposure. Since TDG-deficient cells have not yet been reported [31], it is impossible to state with certainty whether this glycosylase influences the cellular response to 5-FU.
3.2 Downstream components of BER
Few studies to date have looked at the involvement of BER components downstream of the DNA glycosylases with regards to 5-FU sensitivity. APE1 is the major abasic endonuclease in mammalian cells, and operates centrally in the BER response after glycosylase-catalyzed base release (Figure 3). APE1 appears to be essential for mammalian cell viability [47,48], yet expressing a dominant-negative APE1 variant (termed ED), which binds with high affinity to substrate DNA and blocks subsequent repair steps, was found to recapitulate the cellular sensitivity to alkylating agents seen with AP endonuclease deficient E. coli and S. cerevisiae [49]. Moreover, recent work has found that ED expression in Chinese hamster ovary (CHO) cells significantly increases sensitivity to 5-FU (∼five-fold) and FdUrd (∼thirty-fold), suggesting the formation of an APE1-specific substrate (presumably an AP site), blockage of the normal repair response, and consequent activation of cell death (McNeill and Wilson, manuscript in preparation).
DNA Polymerase β (POLβ) performs two important biochemical functions in mammalian BER, namely nucleotide gap filling and 5′-dRP excision, which immediately follow AP site incision by APE1 (Figure 3). Surprisingly, studies have found that Pol β −/− MEFs are more resistant to FdUrd (5-FU was not examined) than their wild-type counterparts [50,51]. Furthermore, studies examining CHO cells defective in XRCC1, a protein critical to single strand break (SSB) repair through interactions with POLβ and DNA ligase IIIα, have found no obvious role for this protein in 5-FU [52] or FdUrd resistance (Li and Wyatt, unpublished results). The lack of involvement of POLβ and XRCC1 in 5-FU responsiveness is striking given their prominent role in the later steps of BER. One possible explanation might be altered sub-pathway choice depending on the cell type examined [53]. For example, long-patch BER may complete the steps downstream of APE1 incision, during which POLδ/ε performs nucleotide synthesis, flap endonuclease (FEN-1) removes the nucleotide overhang terminated by the 5′-dRP group, and DNA ligase I seals the nick (Figure 3). Why short-patch BER might be deleterious or dispensable under conditions of TS inhibition and/or 5-FU incorporation is unclear and requires further investigation. Moreover, studies need to more intensively delineate the contribution of 5-FU versus BER intermediates in cell death.
In S. cerevisiae, the influence of several BER components in response to 5-FU has been examined [54], although there are a number of biochemical differences in the BER steps between S. cerevisiae and mammalian cells to appreciate [55]. S. cerevisiae possess a UNG homolog, but lack SMUG1, MBD4, and TDG homologues [56]. S. cerevisiae deficient in UNG were more resistant to 5-FU than the wild-type strain, suggesting that 5-FU (or uracil) excision and generation of repair intermediates is vital to toxicity in this model system [54]. Conversely, a strain deficient in APN1, the major abasic endonuclease of budding yeast, was exquisitely sensitive to 5-FU compared to a wild-type strain [54], implying that the ability to process AP sites in yeast (and mammalian cells, see above) is crucial for 5-FU resistance. S. cerevisiae also lack a paralog of POLβ. It is believed that the 5′-dRP group is instead removed by the 5′-flap endonuclease RAD27 (or FEN1 in mammals) as part of a displaced strand, similar in design to the FEN1-dependent long-patch BER carried out in mammalian cells (Figure 3). Intriguingly, a rad27 null strain of S. cerevisiae was extremely resistant to 5-FU [54], which parallels the observation that Pol β -/- MEFs are resistant to FdUrd [50]. Collectively, the results examining 5-FU and BER components in S. cerevisiae generally mirror those seen in mammalian cells, but such comparisons must be carefully made given the differences in cell death processes between these disparate species.
3.3 BER futile cycling during treatment with TS inhibitors
One of the more interesting aspects regarding the role of BER during TS inhibition is the notion that the repair process acts as an unwitting executioner [57]. Recall that suicide inhibition of TS by FdUMP causes an increase in dUTP, which can become incorporated into DNA during replication. Because BER requires a DNA resynthesis step following uracil excision, elevated dUTP presumably causes reintroduction of uracil into DNA to create a ‘futile cycling’ of attempted repair [58]. Unrepaired BER intermediates, namely abasic sites and SSBs, are known to be toxic and clastogenic DNA lesions, reviewed in [59]. Thus, repetitive uracil excision during TS inhibition is thought to contribute to cellular lethality.
The evidence in favor of BER futile cycling stems in large part from studies examining the crucial roles of dUTPase and dUTP levels in mediating toxicity caused by TS inhibitors. Several studies have established a direct relationship between dUTP pools, the extent of DNA fragmentation, and cytotoxicity [60-64]. DNA strand breaks were measured by pulsed-field electrophoresis or the comet assay, yet these investigations did not explicitly differentiate between SSBs and double strand breaks (DSBs). Following on from the observations of elevated intracellular dUTP levels affecting DNA integrity and cellular viability, a number of studies specifically modulated the levels of dUTPase [65-68]. Interestingly, increasing dUTPase activity only delayed, but did not prevent the lethality of TS inhibitors, implying that cell death does not entirely depend on DNA damage resulting from uracil incorporation [67,68]. In total, the early studies established important associations between TS inhibition, the formation of DNA strand breaks and lethality, but left unanswered important questions regarding which specific DNA repair pathways and proteins might contribute to the formation or resolution of the strand breaks.
The source of strand breaks observed following treatment with TS inhibitors in mammalian cells was proposed to be BER-mediated, although not experimentally demonstrated to be dependent on specific BER components. BER generates a SSB intermediate, so additional events would be required to create a DSB following uracil incorporation and excision from the daughter strand during replication. In one series of studies, an endonuclease activity was implicated in the production of DNA strand breaks following TS inhibition [69,70]. However, it was not established whether the endonuclease activity was associated with DNA repair or the execution of apoptosis. In addition, the studies that examined DNA strand breaks utilized anti-folates (e.g. methotrexate, CB3717, raltitrexed) or FdUrd, not 5-FU. Thus, the contribution of genomic 5-FU (its incorporation or excision) was not explicitly examined.
Caradonna and coworkers reported that preventing BER futile cycling from occurring confers resistance to FdUrd [71]. In brief, they found that resistance to FdUrd is observed in certain human cell lines in which the nuclear isoform of UNG (UNG2) is prematurely degraded in S-phase following FdUrd treatment, as opposed to G2 when UNG2 is normally degraded [71]. They speculated that this premature degradation confers resistance to FdUrd by preventing the futile BER response. However, there has been a question regarding the interpretation of the FACS analysis used to conclude that the proposed premature UNG2 degradation actually occurred in S-phase [72]. Nonetheless, Fischer et al. showed at least in HeLa cells that siRNA against UNG2 conferred resistance to FdUrd [71], offering evidence in support of the futile cycling model.
The prediction of the BER futile cycling model is that restraining UDG-initiated BER would protect against the toxicity of TS inhibition. However, prior studies with the Ung -/- MEFs showed no differential sensitivity to 5-FU or FdUrd (see Section 3.1). Interestingly, these experiments also showed an accumulation of genomic uracil, implying that the accumulation of downstream BER intermediates is responsible for the cytotoxicity of fluoropyrimidines. To our knowledge, no studies have simultaneously manipulated UDG and dUTPase activity in mammalian cells to test whether tolerance of genomic uracil occurs as a means of developing resistance to TS inhibitors. The ‘tolerance’ of genomic 5-FU is a topic touched upon both above and in Section 4.
4. Mismatch Repair and S-phase Checkpoint Signaling pathways
4.1 Mismatch Repair
MMR is responsible for correcting replication errors such as base:base mismatches and polymerase slippage products (i.e. insertion/deletion loops) at nucleotide repeat sequences [56]. As noted above, germ-line mutations in MMR genes have been found to give rise to HNPCC, thus linking a specific repair defect with predisposition to colorectal and other cancers [43]. Biochemically, base:base mismatches are recognized by a heterodimeric protein complex (MSH2:MSH6, also known as MutSα). The second step of MMR involves recognition of the bound MutSα by a second heterodimer (MLH1:PMS2, also known as MutLα). Recruitment of MutLα signals exonucleases to degrade the daughter strand containing the mismatch, and subsequently polymerase and ligase activities to complete repair (Figure 4). Mutations in MLH1 and MSH2 seem to account for the majority of HNPCC cases [43], while silencing of MLH1 by promoter hypermethylation is a frequent event in sporadic colorectal cancer with MSI-H [73,74]. In addition to correcting replication errors, MMR also plays an important role in apoptotic signaling in response to DNA damage [75-77]. Specifically, MMR recognition of damaged DNA can signal to the cell death machinery to trigger apoptosis, so that loss of MMR by genetic or epigenetic means can promote a ‘tolerance’ to DNA damage and resistance to chemotherapeutic DNA damaging agents.
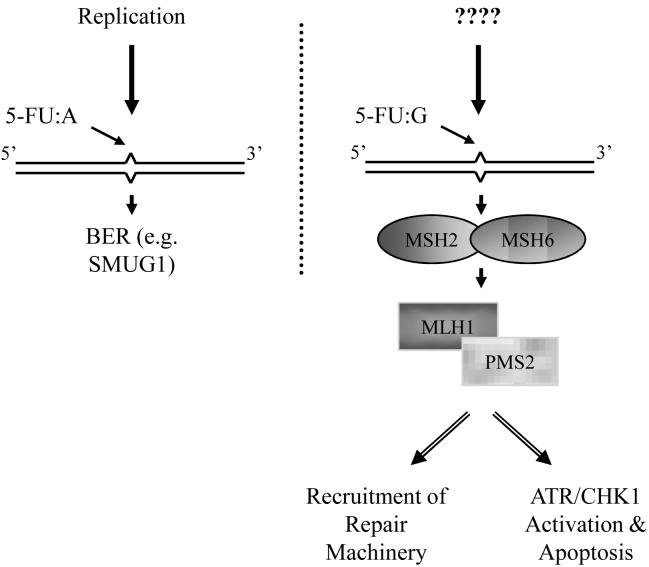
Figure 4. MMR response to genomic 5-FU.
5-FU:A pairs that arise upon incorporation of FdUTP into the genome during DNA synthesis are recognized and processed by the BER pathway (left; see text and Figure 3 for details). 5-FU:G pairs, however, are recognized by the MMR MutSa complex comprised of MSH2:MHS6 (right). Subsequently, the MutLα heterdimer made up of MLH1:PMS2 associates and either initiates a repair response or triggers apoptotic signaling through ATR/CHK1 activation. A burning question that remains though is “how do 5-FU:G mispairs arise in chromosomal DNA?” See text for further details.
It has been demonstrated in biochemical assays that human MutSα can recognize 5-FU paired opposite guanine but not adenine in DNA [36,78], and 5-FU:G mispairs are efficiently corrected in MMR proficient cell extracts [36]. In a similar vein, MutSα recognizes U:G but not U:A pairs in DNA [78]. Thus, MMR recognition of 5-FU may not be recognition per se, but detection of a “mismatch”, i.e. a uracil analog paired opposite guanine. The hMSH2:hMSH3 complex (MutSβ), which recognizes insertion/deletion loops, does not recognize 5-FU opposite adenine or guanine [78].
A number of studies have reported that cells deficient in MMR components, particularly MSH2 and MLH1, are resistant to 5-FU [78-80], which fits with the model that MMR-dependent recognition of certain forms of DNA damage initiates apoptosis. Genomic 5-FU paired opposite guanine was higher in MMR-deficient cells, implying that the presence of 5-FU was being tolerated due to the loss of MMR [78]. This is parallel to the phenomenon of apoptotic cell death being induced by an MMR-dependent recognition of O6-methylguanine opposite thymine [59]. However, there is an aspect of the studies of MMR and genomic 5-FU that is ambiguous. Specifically, how do 5-FU:G mispairs occur in chromosomal DNA? U:A pairs arise when dUTP is incorporated by DNA polymerases, while U:G mispairs occur through cytosine deamination [56]. MMR-dependent cell cycle arrest following 5-FU treatment has been found to take place in the first cell cycle [78,81], seemingly requiring that a DNA polymerase insert 5-FU opposite a guanine. However, from the limited biochemical evidence available, FdUTP is incorporated opposite adenine. Future studies aimed at delineating the mechanism of 5-FU:G formation are necessary, perhaps examining whether one of the various DNA polymerases has the capacity to insert 5-FU opposite guanine.
It is important to point out that, depending on the cell model, the MMR-dependent influence on 5-FU toxicity is determined by the duration and dose of the base analog [36,81], suggesting that other consequences of 5-FU treatment contribute to the cell death response. BER status and dUTPase activity were not evaluated in any of the above MMR models, so it is unclear to what extent MMR and BER collectively contribute to 5-FU cellular sensitivity. Similarly, clinical studies examining 5-FU response in MSI-H patients do not provide a clear picture of the specific involvement of MMR. In particular, an early report offered promise that MSI-H patients might selectively benefit from 5-FU treatment [82]. However, other studies have since found that patients with tumors lacking MSI (i.e., MMR proficient) more significantly benefit from 5-FU treatment [83-85], while other studies report no obvious difference in 5-FU response and MSI status [86,87]. BER status was not simultaneously examined in any of these clinical studies.
4.2 Checkpoint Signaling and other repair pathways
There is growing momentum in targeting DNA damage and cell cycle checkpoint signaling pathways as a means of cancer treatment [88,89]. This is relevant for 5-FU therapy, as TS inhibition and incorporation of the fluorinated base into DNA occurs during S-phase. The PI3K-like kinases, ATM (ataxia telangiectasia-mutated) and ATR (ATM-related), are central mediators in the response to DNA damage during S-phase [90], and their protein substrates number over 700 [91]. CHK1 is thought to be an important downstream target of ATR and is phosphorylated by ATR in response to replication stress [90]. There is some evidence suggesting that S-phase checkpoint pathways respond to 5-FU treatment and TS inhibition. For instance, ATR hypomorphic cells are hypersensitive to 5-FU [92]. Furthermore, TS inhibitors induce CHK1 phosphorylation [93] and phospho-CHK1 foci that colocalize with replication protein A [94]. UCN-01 inhibits the CHK1 kinase and has reached clinical trials [89]. Notably, coadministration of UCN-01 with 5-FU increases sensitivity [95], and Chk1 deficiency similarly sensitizes cells to 5-FU [96,97]. A recent report suggests that ATR and CHK1 status influence cellular sensitivity to 5-FU in a manner that is dependent on MMR- or BER-mediated responses, dictated by the drug dose and exposure period [81].
Nucleotide excision repair copes with bulky helix-distorting lesions, such as those generated by ultraviolet light or the crosslinking agent cisplatin [56]. Although it seems unlikely that nucleotide excision repair would participate in a 5-FU response, given its preference for larger DNA adducts, there is evidence that the pathway recognizes and excises more subtle base lesions, such as 8-oxoguanine [98]. To our knowledge, the contribution of NER to 5-FU resistance has not been explored. Repair pathways for DNA DSBs are worth mentioning because of the studies cited earlier that identified associations between strand breaks and the lethality of TS inhibitors. There are two major repair pathways that respond to DNA DSBs, namely homologous recombination (HR) and non-homologous end-joining (NHEJ). CHO cell lines defective in components of NHEJ (Ku80 and DNA-PKcs), HR (XRCC3, a RAD51 family member), and S-phase checkpoint signaling (XRCC8, an ataxia telangiectasia-like mutant) do not appear to be specifically sensitive to 5-FU treatment [52]. Nonetheless, there appears to be a number of reasons to examine HR more carefully. First, transient depletion of the RAD51 recombinase by siRNA rendered cells sensitive to thymidylate deprivation induced by an anti-folate TS inhibitor [94]. Second, at least for alkylation damage, HR seems to resolve aberrant DNA structures (e.g. DSBs that arise during replication in S-phase) caused by both unrepaired BER intermediates and MMR-dependent recognition of O6-methylguanine opposite thymine [59]. Considering the evidence for BER and MMR involvement in 5-FU management (see Sections 3 and 4.1), it is reasonable to presume that HR would be involved in a compensatory response to 5-FU-related DNA damage. Third, there are interesting links among S-phase checkpoint signaling and the HR machinery. For example, CHK1 has been reported to be required for HR [99]. Thus, the observed sensitization of cells to 5-FU when CHK1 is deficient also suggests that HR may be required.
Closing Thoughts
5-FU and its pro-drug derivative capecitabine are commonly employed today in the clinic to eradicate or manage various solid tumors, most notably of the colon. Evidence clearly indicates that products of 5-FU metabolism can affect intracellular nucleotide pools, and ultimately lead to the incorporation of “false” bases, i.e. uracil and 5-FU, into genomic DNA. Thus, it is not surprising that recent studies have suggested that DNA damage responses play a key role in dictating cellular responsiveness to 5-FU exposure.
To date, the pathways that appear most relevant in determining 5-FU outcome are BER and MMR (summarized in Table 1). The finding that the latter pathway contributes to 5-FU sensitivity is striking, given that genetic mutations that disrupt MMR capacity predispose for colorectal cancer. As studies are mixed regarding the clinical efficacy of 5-FU treatment as predicted by MMR genotype, it stands to reason that other factors play at minimum equally vital roles in determining individual responsiveness to 5-FU exposure. One such pathway is undoubtedly BER (namely the proteins SMUG1 and the abasic endonuclease, see Section 3), although mechanistic details need to be elucidated. In particular, the relative contribution of genomic uracil, genomic 5-FU, abasic sites, and strand break intermediates to cytotoxicity remains unclear. Furthermore, DNA damage responses including HR have yet to be extensively interrogated (Table 1). While past investigations have largely focused on measuring TS (target), TK (activator) and dihydropyrimidine dehydrogenase (breaks down 5-FU) for predicting tumor response, we suggest that future studies focus on repair potential, e.g. deleterious polymorphisms in BER genes, as markers for forecasting 5-FU outcome.
Table 1.
Major DNA Repair Processes and Projected Involvement in 5-FU Response.
| DNA Repair Pathway | Primary DNA Substrates | Anticipated Involvement |
|---|---|---|
| MGMT = O6-methylguanine DNA methyltransferase | ||
| ABH = AlkB homolog dioxygenase | ||
| Direct Reversal | None | |
| MGMT | O6-alkylguanine | |
| ABH family | N1-alkylpurines | |
| N3-alkylpyrimidines | ||
| BER | Small base modifications, abasic sites, SSBs | See Section 3 |
| MMR | Base-base mismatches and small insertion/deletion loops | See Section 4.1 |
| NER | Helix-distorting base adducts | untested, unlikely |
| Recombination | ||
| NHEJ | DSBs | Not likely |
| HR | DSBs and collapsed replication forks | See Section 4.2 |
An emerging interest in the field of DNA repair is the prospect of manipulating damage response systems to either augment cellular resistance (improve repair) or increase cellular sensitivity (inhibit repair) to enhance therapeutic efficacy of the many DNA damaging drugs used in the clinic. Such agents typically cause different types of DNA modifications, thus potentially invoking several DNA repair mechanisms. In this regard, 5-FU is no different (Table 1). As the factors that are most critical in determining 5-FU responsiveness become identified, novel agents can be designed to selectively inactivate or enhance these key components. Future combination therapies can thus be designed with a better knowledge of which DNA repair and signaling response(s) is invoked and should be targeted. Such information will become crucial as new generations of inhibitors enter the clinic and are used in combination therapies with the established chemotherapeutics that damage DNA.
Acknowledgements
This article was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (DMW), and by NIH grant 1 R01 CA100450 (MDW). The authors would like to extend special thanks to their family members, “Coach” and “Grandpa” Parks.
Abbreviations
- 5-FU
5-fluorouracil
- FdUrd
5-fluoro-2′-deoxyuridine
- TS
thymidylate synthase
- TMP
thymidylate
- TTP
thymidine triphosphate
- dUMP
deoxyuridylate
- dUTP
deoxyuridine triphosphate
- TK
thymidine kinase
- dUTPase
deoxyuridine triphosphate nucleotidohydrolase
- SSB
single strand break
- DSB
double strand break
- UDG
uracil DNA glycosylase
- BER
base excision repair
- AP site
abasic site (apurinic/apyrimidinic)
- MMR
mismatch repair
- HR
homologous recombination
- CHO
Chinese hamster ovary
References
- 1.Rutman RJ, Cantarow A, Paschkis KE. The catabolism of uracil in vivo and in vitro. J. Biol. Chem. 1954;210:321–9. [PubMed] [Google Scholar]
- 2.Heidelberger C, Leibman KC, Harbers E, Bhargava PM. The comparative utilization of uracil-2-C14 by liver, intestinal mucosa, and Flexner-Jobling carcinoma in the rat. Cancer Res. 1957;17:399–404. [PubMed] [Google Scholar]
- 3.Duschinsky R, Pleven E, Heidelberger C. The synthesis of 5-fluoropyrimidines. J. Am. Chem. Soc. 1957;79:4559–4560. [Google Scholar]
- 4.Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, Schnitzer RJ, Pleven E, Scheiner J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–6. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 5.Holland JF, Frei E. Cancer Medicine 7. BC Decker; Hamilton, Ontario: 2006. [Google Scholar]
- 6.Weisberg E, Manley PW, Cowan-Jacob SW, Hochhaus A, Griffin JD. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat. Rev. Cancer. 2007;7:345–56. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 7.Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26:3637–43. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Josef E. Capecitabine and radiotherapy as neoadjuvant treatment for rectal cancer. Am. J. Clin. Oncol. 2007;30:649–55. doi: 10.1097/COC.0b013e3180ca7c9e. [DOI] [PubMed] [Google Scholar]
- 9.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 10.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 1990;48:381–95. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 11.el-Hajj HH, Wang L, Weiss B. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 1992;174:4450–4456. doi: 10.1128/jb.174.13.4450-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425–31. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg JM, Tymoczko JL, Stryer L. Biochemistry. W. H. Freeman; New York, NY: 2007. [Google Scholar]
- 14.Berger FG, Berger SH. Thymidylate synthase as a chemotherapeutic drug target: where are we after fifty years? Cancer Biol. Ther. 2006;5:1238–41. doi: 10.4161/cbt.5.9.3414. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Yoshida S, Saneyoshi M, Yamaguchi T. Utilization of 5-fluoro-2′-deoxyuridine triphosphate and 5-fluoro-2′-deoxycytidine triphosphate in DNA synthesis by DNA polymerases alpha and beta from calf thymus. Cancer Res. 1981;41:4132–5. [PubMed] [Google Scholar]
- 16.Caradonna SJ, Cheng YC. The role of deoxyuridine triphosphate nucleotidohydrolase, uracil-DNA glycosylase, and DNA polymerase alpha in the metabolism of FUdR in human tumor cells. Mol. Pharmacol. 1980;18:513–20. [PubMed] [Google Scholar]
- 17.Cheng YC, Nakayama K. Effects of 5-fluoro-2′-deoxyuridine on DNA metabolism in HeLa cells. Mol. Pharmacol. 1983;23:171–4. [PubMed] [Google Scholar]
- 18.Danenberg PV, Heidelberger C, Mulkins MA, Peterson AR. The incorporation of 5-fluoro-2′-deoxyuridine into DNA of mammalian tumor cells. Biochem. Biophys. Res. Commun. 1981;102:654–8. doi: 10.1016/s0006-291x(81)80182-9. [DOI] [PubMed] [Google Scholar]
- 19.Ingraham HA, Tseng BY, Goulian M. Nucleotide levels and incorporation of 5-fluorouracil and uracil into DNA of cells treated with 5-fluorodeoxyuridine. Mol. Pharmacol. 1982;21:211–6. [PubMed] [Google Scholar]
- 20.Kufe DW, Major PP, Egan EM, Loh E. 5-Fluoro-2′-deoxyuridine incorporation in L1210 DNA. J. Biol. Chem. 1981;256:8885–8. [PubMed] [Google Scholar]
- 21.Kufe DW, Scott P, Fram R, Major P. Biologic effect of 5-fluoro-2′-deoxyuridine incorporation in L1210 deoxyribonucleic acid. Biochem. Pharmacol. 1983;32:1337–40. doi: 10.1016/0006-2952(83)90443-4. [DOI] [PubMed] [Google Scholar]
- 22.Lonn U, Lonn S. DNA lesions in human neoplastic cells and cytotoxicity of 5-fluoropyrimidines. Cancer Res. 1986;46:3866–70. [PubMed] [Google Scholar]
- 23.Major PP, Egan E, Herrick D, Kufe DW. 5-Fluorouracil incorporation in DNA of human breast carcinoma cells. Cancer Res. 1982;42:3005–9. [PubMed] [Google Scholar]
- 24.Parker WB, Kennedy KA, Klubes P. Dissociation of 5-fluorouracil-induced DNA fragmentation from either its incorporation into DNA or its cytotoxicity in murine T-lymphoma (S-49) cells. Cancer Res. 1987;47:979–82. [PubMed] [Google Scholar]
- 25.Sawyer RC, Stolfi RL, Martin DS, Spiegelman S. Incorporation of 5-fluorouracil into murine bone marrow DNA in vivo. Cancer Res. 1984;44:1847–51. [PubMed] [Google Scholar]
- 26.Tanaka M, Kimura K, Yoshida S. Enhancement of the incorporation of 5-fluorodeoxyuridylate into DNA of HL-60 cells by metabolic modulations. Cancer Res. 1983;43:5145–50. [PubMed] [Google Scholar]
- 27.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–59. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 30.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–36. doi: 10.1074/jbc.M207107200. Epub 2002 Aug 02. [DOI] [PubMed] [Google Scholar]
- 31.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Ingraham HA, Tseng BY, Goulian M. Mechanism for exclusion of 5-fluorouracil from DNA. Cancer Res. 1980;40:998–1001. [PubMed] [Google Scholar]
- 33.Mauro DJ, De Riel JK, Tallarida RJ, Sirover MA. Mechanisms of excision of 5-fluorouracil by uracil DNA glycosylase in normal human cells. Mol. Pharmacol. 1993;43:854–7. [PubMed] [Google Scholar]
- 34.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J. Biol. Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 35.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 36.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–68. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 37.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–5. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 38.Andersen S, Heine T, Sneve R, König I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 39.Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair (Amst) 2008;7:162–169. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortellino S, Turner D, Masciullo V, Schepis F, Albino D, Daniel R, Skalka AM, Meropol NJ, Alberti C, Larue L, Bellacosa A. The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity. Proc. Natl. Acad. Sci. USA. 2003;100:15071–15076. doi: 10.1073/pnas.2334585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansom OJ, Zabkiewicz J, Bishop SM, Guy J, Bird A, Clarke AR. MBD4 deficiency reduces the apoptotic response to DNA-damaging agents in the murine small intestine. Oncogene. 2003;22:7130–6. doi: 10.1038/sj.onc.1206850. [DOI] [PubMed] [Google Scholar]
- 42.Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, Golemis EA, Genuardi M, Neri G. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc. Natl. Acad. Sci. USA. 1999;96:3969–74. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiricny J, Nystrom-Lahti M. Mismatch repair defects in cancer. Curr. Opin. Genet. Dev. 2000;10:157–61. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 44.Bader S, Walker M, Harrison D. Most microsatellite unstable sporadic colorectal carcinomas carry MBD4 mutations. Br. J. Cancer. 2000;83:1646–9. doi: 10.1054/bjoc.2000.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bader S, Walker M, Hendrich B, Bird A, Bird C, Hooper M, Wyllie A. Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency. Oncogene. 1999;18:8044–7. doi: 10.1038/sj.onc.1203229. [DOI] [PubMed] [Google Scholar]
- 46.Riccio A, Aaltonen LA, Godwin AK, Loukola A, Percesepe A, Salovaara R, Masciullo V, Genuardi M, Paravatou-Petsotas M, Bassi DE, Ruggeri BA, Klein-Szanto AJ, Testa JR, Neri G, Bellacosa A. The DNA repair gene MBD4 (MED1) is mutated in human carcinomas with microsatellite instability. Nat. Genet. 1999;23:266–8. doi: 10.1038/15443. [DOI] [PubMed] [Google Scholar]
- 47.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–70. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 48.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. USA. 2005;102:5739–43. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNeill DR, Wilson DM., 3rd A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol. Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Berger SH, Wyatt MD. Involvement of base excision repair in response to therapy targeted at thymidylate synthase. Mol. Cancer Ther. 2004;3:747–753. [PubMed] [Google Scholar]
- 51.Li L, Connor EE, Berger SH, Wyatt MD. Determination of apoptosis, uracil incorporation, DNA strand breaks, and sister chromatid exchanges under conditions of thymidylate deprivation in a model of BER deficiency. Biochem. Pharmacol. 2005;70:1458–1468. doi: 10.1016/j.bcp.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Haveman J, Castro Kreder N, Rodermond HM, van Bree C, Franken NA, Stalpers LJ, Zdzienicka MZ, Peters GJ. Cellular response of X-ray sensitive hamster mutant cell lines to gemcitabine, cisplatin and 5-fluorouracil. Oncol. Rep. 2004;12:187–92. doi: 10.3892/or.12.1.187. [DOI] [PubMed] [Google Scholar]
- 53.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seiple L, Jaruga P, Dizdaroglu M, Stivers JT. Linking uracil base excision repair and 5-fluorouracil toxicity in yeast. Nucleic Acids Res. 2006;34:140–51. doi: 10.1093/nar/gkj430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelley MR, Kow YW, Wilson DM., 3rd Disparity between DNA base excision repair in yeast and mammals: translational implications. Cancer Res. 2003;63:549–54. [PubMed] [Google Scholar]
- 56.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D.C: 2006. [Google Scholar]
- 57.Berger SH, Pittman DL, Wyatt MD. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem. Pharmacol. 2008;67:697–706. doi: 10.1016/j.bcp.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aherne GW, Brown S. The role of uracil misincroporation in thymineless death. In: Jackman AL, editor. Anticancer Drug Development Guide: Antifolate Drugs in Cancer Therapy. Humana Press Inc; Totowa, NJ: 1999. pp. 409–421. [Google Scholar]
- 59.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–94. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canman CE, Lawrence TS, Shewach DS, Tang HY, Maybaum J. Resistance to fluorodeoxyuridine-induced DNA damage and cytotoxicity correlates with an elevation of deoxyuridine triphosphatase activity and failure to accumulate deoxyuridine triphosphate. Cancer Res. 1993;53:5219–24. [PubMed] [Google Scholar]
- 61.Canman CE, Tang HY, Normolle DP, Lawrence TS, Maybaum J. Variations in patterns of DNA damage induced in human colorectal tumor cells by 5-fluorodeoxyuridine: implications for mechanisms of resistance and cytotoxicity. Proc. Natl. Acad. Sci. USA. 1992;89:10474–10478. doi: 10.1073/pnas.89.21.10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curtin NJ, Harris AL, Aherne GW. Mechanism of cell death following thymidylate synthase inhibition: 2′-deoxyuridine-5′-triphosphate accumulation, DNA damage, and growth inhibition following exposure to CB3717 and dipyridamole. Cancer Res. 1991;51:2346–52. [PubMed] [Google Scholar]
- 63.Ingraham HA, Dickey L, Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986;25:3225–3230. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- 64.Sedwick WD, Kutler M, Brown OE. Antifolate-induced misincorporation of deoxyuridine monophosphate into DNA: inhibition of high molecular weight DNA synthesis in human lymphoblastoid cells. Proc. Natl. Acad. Sci. USA. 1981;78:917–921. doi: 10.1073/pnas.78.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Canman CE, Radany EH, Parsels LA, Davis MA, Lawrence TS, Maybaum J. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 1994;54:2296–2298. [PubMed] [Google Scholar]
- 66.Koehler SE, Ladner RD. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol. Pharmacol. 2004;66:620–626. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 67.Parsels LA, Parsels JD, Wagner LM, Loney TL, Radany EH, Maybaum J. Mechanism and pharmacological specificity of dUTPase-mediated protection from DNA damage and cytotoxicity in human tumor cells. Cancer Chemother. Pharmacol. 1998;42:357–362. doi: 10.1007/s002800050829. [DOI] [PubMed] [Google Scholar]
- 68.Webley SD, Welsh SJ, Jackman AL, Aherne GW. The ability to accumulate deoxyuridine triphosphate and cellular response to thymidylate synthase (TS) inhibition. Br. J. Cancer. 2001;85:446–452. doi: 10.1054/bjoc.2001.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshioka A, Tanaka S, Hiraoka O, Koyama Y, Hirota Y, Ayusawa D, Seno T, Garrett C, Wataya Y. Deoxyribonucleoside triphosphate imbalance. 5-Fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J. Biol. Chem. 1987;262:8235–8241. [PubMed] [Google Scholar]
- 70.Ayusawa D, Arai H, Wataya Y, Seno T. A specialized form of chromosomal DNA degradation induced by thymidylate stress in mouse FM3A cells. Mutat. Res. 1988;200:221–30. doi: 10.1016/0027-5107(88)90086-3. [DOI] [PubMed] [Google Scholar]
- 71.Fischer JA, Muller-Weeks S, Caradonna SJ. Fluorodeoxyuridine Modulates Cellular Expression of the DNA Base Excision Repair Enzyme Uracil-DNA Glycosylase. Cancer Res. 2006;66:8829–37. doi: 10.1158/0008-5472.CAN-06-0540. [DOI] [PubMed] [Google Scholar]
- 72.Buchegger F, Dupertuis YM, Perillo-Adamer F. A pitfall of propidium iodide staining in fluorescence-activated cell sorting cell cycle analysis? Cancer Res. 2007;67:5576–7. doi: 10.1158/0008-5472.CAN-06-3663. author reply 5577. [DOI] [PubMed] [Google Scholar]
- 73.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–11. [PubMed] [Google Scholar]
- 74.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–8. [PubMed] [Google Scholar]
- 75.Bignami M, O'Driscoll M, Aquilina G, Karran P. Unmasking a killer: DNA O(6)-methylguanine and the cytotoxicity of methylating agents. Mutat. Res. 2000;462:71–82. doi: 10.1016/s1383-5742(00)00016-8. [DOI] [PubMed] [Google Scholar]
- 76.O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–92. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 77.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst) 2004;3:1091–101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 78.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J. Biol. Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 79.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–31. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meyers M, Wagner MW, Hwang HS, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA mismatch repair protein in fluoropyrimidine-mediated cell death and cell cycle responses. Cancer Res. 2001;61:5193–201. [PubMed] [Google Scholar]
- 81.Liu A, Yoshioka KI, Salerno V, Hsieh P. The mismatch repair-mediated cell cycle checkpoint response to fluorodeoxyuridine. J. Cell Biochem. 2008;105:245–54. doi: 10.1002/jcb.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hemminki A, Mecklin JP, Jarvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–8. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 83.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 84.Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Pinol V, Xicola RM, Bujanda L, Rene JM, Clofent J, Bessa X, Morillas JD, Nicolas-Perez D, Paya A, Alenda C. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy in colorectal cancer. Gut. 2006;55:848–55. doi: 10.1136/gut.2005.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 87.Lamberti C, Lundin S, Bogdanow M, Pagenstecher C, Friedrichs N, Buttner R, Sauerbruch T. Microsatellite instability did not predict individual survival of unselected patients with colorectal cancer. Int. J. Colorectal Dis. 2007;22:145–52. doi: 10.1007/s00384-006-0131-8. [DOI] [PubMed] [Google Scholar]
- 88.Ashwell S, Zabludoff S. DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin. Cancer Res. 2008;14:4032–7. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- 89.Tse AN, Carvajal R, Schwartz GK. Targeting checkpoint kinase 1 in cancer therapeutics. Clin. Cancer Res. 2007;13:1955–60. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 90.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 91.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 92.Wilsker D, Bunz F. Loss of ataxia telangiectasia mutated- and Rad3-related function potentiates the effects of chemotherapeutic drugs on cancer cell survival. Mol. Cancer Ther. 2007;6:1406–13. doi: 10.1158/1535-7163.MCT-06-0679. [DOI] [PubMed] [Google Scholar]
- 93.Parsels LA, Parsels JD, Tai DC, Coughlin DJ, Maybaum J. 5-fluoro-2′-deoxyuridine-induced cdc25A accumulation correlates with premature mitotic entry and clonogenic death in human colon cancer cells. Cancer Res. 2004;64:6588–94. doi: 10.1158/0008-5472.CAN-03-3040. [DOI] [PubMed] [Google Scholar]
- 94.Yang Z, Waldman AS, Wyatt MD. DNA damage and homologous recombination signaling induced by thymidylate deprivation. Biochem. Pharmacol. 2008;76:987–996. doi: 10.1016/j.bcp.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsueh CT, Kelsen D, Schwartz GK. UCN-01 suppresses thymidylate synthase gene expression and enhances 5-fluorouracil-induced apoptosis in a sequence-dependent manner. Clin. Cancer Res. 1998;4:2201–6. [PubMed] [Google Scholar]
- 96.Robinson HM, Jones R, Walker M, Zachos G, Brown R, Cassidy J, Gillespie DA. Chk1-dependent slowing of S-phase progression protects DT40 B-lymphoma cells against killing by the nucleoside analogue 5-fluorouracil. Oncogene. 2006;25:5359–69. doi: 10.1038/sj.onc.1209532. [DOI] [PubMed] [Google Scholar]
- 97.Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Mol. Cancer Ther. 2006;5:1935–43. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- 98.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA. 1997;94:9463–8. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]