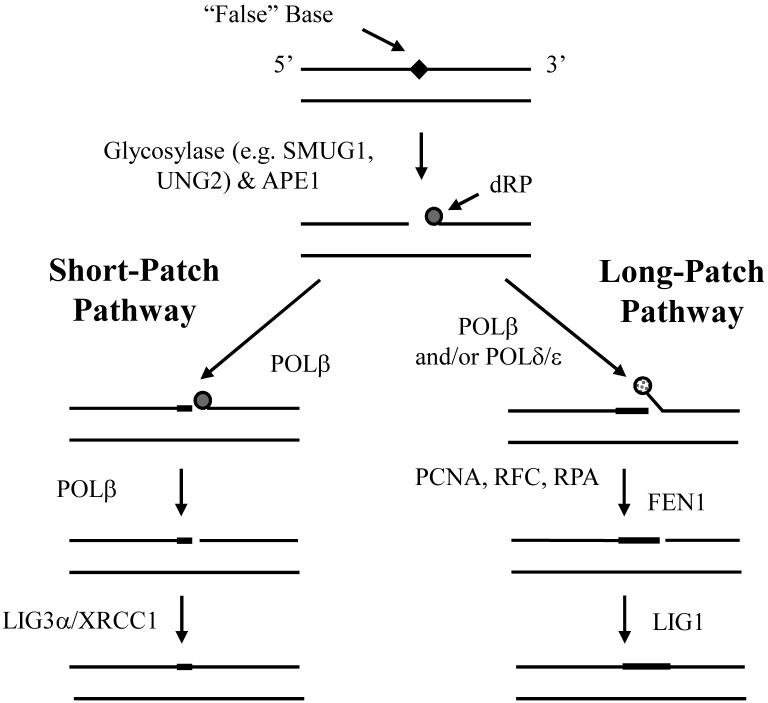
Figure 3. The major enzymatic steps and proteins of mammalian BER.
BER is typically initiated by the removal of an inappropriate (e.g. uracil, 8-oxoguanine or certain mismatches) or “false” base, such as 5-fluorouracil, by a lesion specific DNA glycosylase. Following base excision, the resulting abasic site is most often incised by the major AP endonuclease, APE1, to create a strand break with a 5′-deoxyribose phosphate (dRP) residue. At this point, depending on the nature of the 5′-terminal end and other factors (reviewed in [27]), the DNA gap is restored via either short-patch (left) or long-patch (right) BER. In the former situation, the 5′-dRP residue is excised by the lyase activity of DNA POLb and the single nucleotide gap is filled by the same enzyme. Subsequently, a complex of XRCC1 and DNA ligase IIIα (LIG3) seals the remaining nick. In the case of long-patch BER, the 5′-terminal blocking fragment is ultimately removed by a flap endonuclease (FEN1) following strand-displacement synthesis by POLb and/or POLd/e. After excision of the flap DNA structure, the nick is sealed by DNA LIG1. PCNA, RFC and RPA help facilitate the long-patch repair response. See text for additional details.