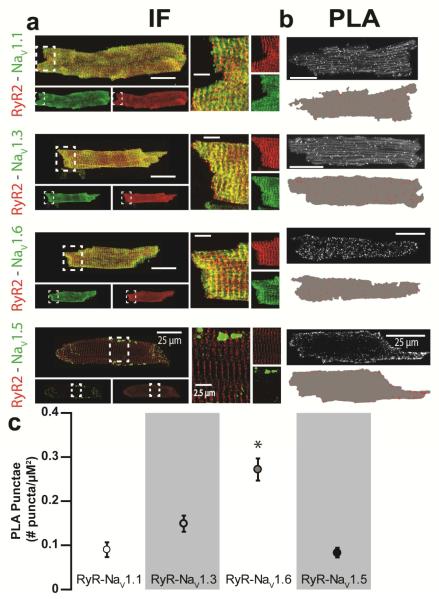
Fig. 4. Neuronal Na+ channels and RyR2 colocalize to the same discrete subcellular regions.
(a) Representative confocal micrographs of isolated CPVT ventricular myocytes labeled for RyR2 (red) with various Nav isoforms (Nav1.x; green) often resulted in an overlap between the immunofluorescent (IF) signals (yellow) when overlaid. Panels on the right show close up views of regions highlighted by dashed white boxes. (b) Representative confocal micrographs of ventricular myocytes isolated from CPVT mice showing fluorescent proximity ligation assay (PLA) signal for RyR2 with different nNav isoforms (NaV1.x). Below each image, are shown the results of digital segmentation with the cell mask in grey and PLA signal in red. (c) Plot of average number of PLA punctae per μm2 (p < 0.001 Kruskal-Wallis test; *, p=0.002, p=0.019, p<0.001 Wilcoxon rank-sum test for Nav1.6 vs. Nav1.1, 1.3 and 1.5, respectively; n = 1231, 1223, 1291 and 2848 punctae from 7, 6, 12 and 7 cells for Nav1.1, 1.3, 1.5 and 1.6, respectively).