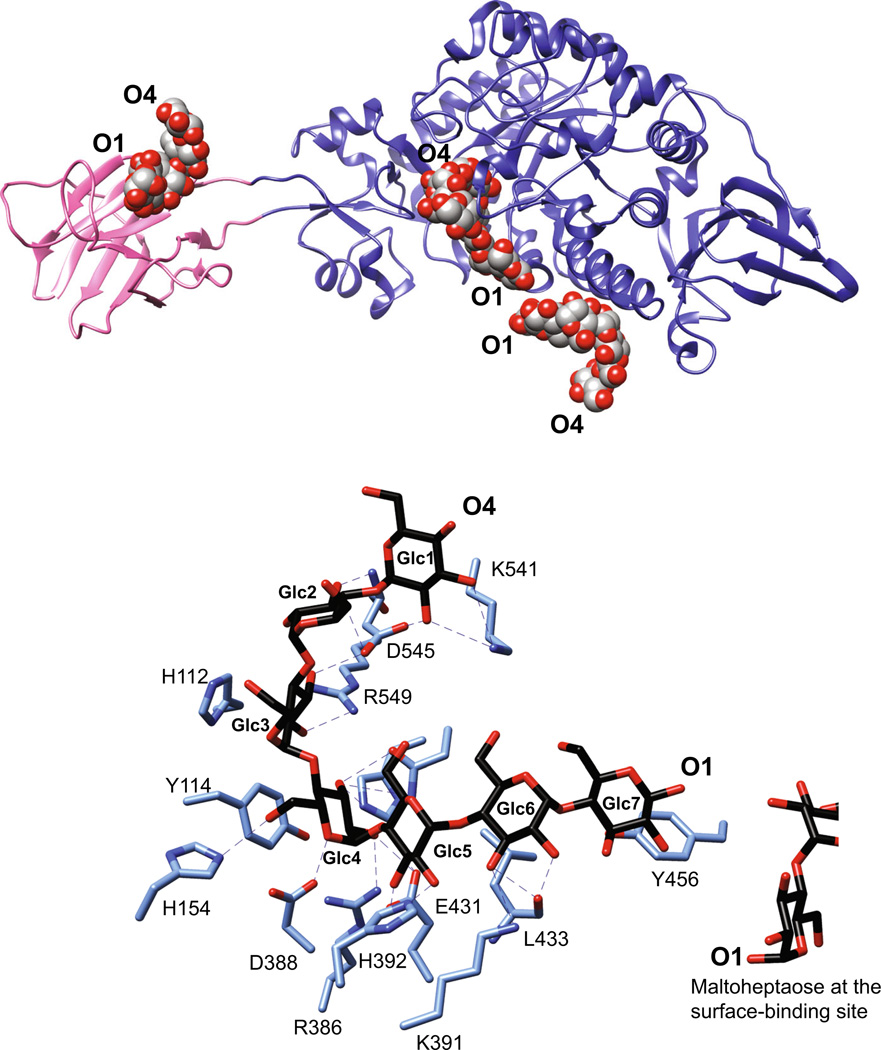
Fig. 3.
SusG is an amylase with a unique CBM insertion. a Structure of the catalytically inactive mutant of SusG D498 N (PDB 3K8L) with bound maltoheptaose. CBM58 (residues 216–335) is highlighted in pink, and maltooligosaccharides bound at CBM58, the active site, and the surface starch-binding site are depicted as spheres. The orientation of the oligosaccharide from the nonreducing end (O4) to reducing end (O1) is indicated. b Close-up view of the active site in the catalytically inactive mutant of SusG D498 N (PDB 3K8L) with bound maltoheptaose. Hydrogen-bonding interactions (≤3.5 Å) are depicted as dashed lines, and Glc residues are labeled from the non-reducing to reducing end