Abstract
Purpose
Lobular carcinoma in situ (LCIS) has been proposed as a non-obligate precursor of invasive lobular carcinoma (ILC). Here we sought to define the repertoire of somatic genetic alterations in pure LCIS and in synchronous LCIS and ILC using targeted massively parallel sequencing.
Methods
DNA samples extracted from microdissected LCIS, ILC and matched normal breast tissue or peripheral blood from 30 patients were subjected to massively parallel sequencing targeting all exons of 273 genes, including the genes most frequently mutated in breast cancer and DNA repair-related genes. Single nucleotide variants and insertions and deletions were identified using state-of-the-art bioinformatics approaches.
Results
The constellation of somatic mutations found in LCIS (n=34) and ILC (n=21) were similar, with the most frequently mutated genes being CDH1 (56% and 66%, respectively), PIK3CA (41% and 52%, respectively) and CBFB (12% and 19%, respectively). Among 19 LCIS and ILC synchronous pairs, 14 (74%) had at least one identical mutation in common, including identical PIK3CA and CDH1 mutations. Paired analysis of independent foci of LCIS from 3 breasts revealed at least one common mutation in each of the 3 pairs (CDH1, PIK3CA, CBFB and PKHD1L1).
Conclusion
LCIS and ILC have a similar repertoire of somatic mutations, with PIK3CA and CDH1 being the most frequently mutated genes. The presence of identical mutations between LCIS-LCIS and LCIS-ILC pairs demonstrates that LCIS is a clonal neoplastic lesion, and provides additional evidence that at least some LCIS are non-obligate precursors of ILC.
Keywords: massively parallel sequencing, invasive lobular carcinoma, lobular carcinoma in situ, clonality, somatic genetic alterations
1. INTRODUCTION
Lobular carcinoma in situ (LCIS) is a non-invasive neoplastic lesion of the breast characterized by expansion of the lobular units by a monomorphic population of dyscohesive cells. Most commonly identified as an incidental finding in otherwise benign breast biopsies, a diagnosis of LCIS confers one of the greatest risks currently recognized for the subsequent development of breast cancer. Yet the biology of this lesion and its potential for progression remain poorly understood. Hence, in clinical practice management, strategies following a diagnosis of LCIS vary widely from observation alone to bilateral prophylactic mastectomy (De Leeuw et al., 1997; Hwang et al., 2004; Lakhani et al., 2006; Vos et al., 1997).
Molecular studies based on the pattern of gene copy number alterations and CDH1 mutations provide supporting evidence that LCIS likely constitutes a non-obligate precursor of invasive lobular carcinoma (ILC) (Andrade et al., 2012; Hwang et al., 2004; Lakhani et al., 1995; Vos et al., 1997). Our group and others have shown that synchronous LCIS and ILC share similar copy number aberrations, predominantly 16q loss and 1q gain (Andrade et al., 2012; Hwang et al., 2004; Lakhani et al., 1995). Historical studies have also demonstrated the presence of the same truncating mutations in the E-cadherin gene CDH1 and loss of heterozygosity (LOH) of the wild-type allele in a small number of synchronous LCIS and ILC cases (Vos et al., 1997). An analysis of 36 ILCs included in the first genomic characterization of breast cancers by The Cancer Genome Atlas (TCGA) revealed that ILCs harbor a repertoire of somatic mutations similar to that of luminal cancers, but have a higher frequency of CDH1 and ERBB2 somatic mutations (Cancer Genome Atlas, 2012; Ciriello G, 2015). To our knowledge, the repertoire of somatic mutations in LCIS has not been reported to date.
The advent of targeted capture massively parallel sequencing has made it possible to investigate somatic mutations from limited amounts of DNA (Wagle et al., 2012). The methodology allows for simultaneous identification of base substitutions, insertions/deletions, copy number aberrations and structural alterations with greater sensitivity and cost effectiveness than traditional sequencing methods. Here we describe the repertoire of genomic changes in fresh-frozen samples of LCIS with or without synchronous ILC subjected to targeted capture massively parallel sequencing of all exons of 273 genes, including the genes most recurrently mutated in breast cancer and genes related to DNA repair. Using this approach, we sought to define the repertoire of genetic alterations in LCIS and ILC, and to investigate the clonal relatedness of synchronous LCIS and ILC, and of multiple foci of LCIS.
2. MATERIALS AND METHODS
2.1 Patients and samples
Patients with a documented history of LCIS, presenting for prophylactic or therapeutic mastectomy, were identified preoperatively and enrolled in a Memorial Sloan Kettering Cancer Center (MSKCC) institutional review board-approved protocol for the collection of fresh-frozen tissue and genomic analyses (IRB 01–135). Following standard clinical sampling, mastectomy specimens were subject to random sampling, and up to 10 fresh-frozen blocks per quadrant were harvested and stored at −80°C for subsequent analysis. For the purposes of this study, 5µm hematoxylin and eosin (H&E) frozen sections were reviewed by at least two study pathologists (DG, MDB, VPA) to identify blocks with LCIS, with or without invasive cancer. Histologic criteria for LCIS and ILC were those described by the 2012 World Health Organization classification (Lakhani S.R. EIO, 2012). Although some patients included in this study also had ductal lesions (invasive ductal cancer or ductal carcinoma in situ) identified on standard clinical pathology, only classic LCIS and ILC lesions harvested as part of protocol 01–135 were included in this study. Matched germline DNA from peripheral blood or normal breast tissue devoid of any neoplastic cells were available for all cases. All samples were anonymized prior to analysis.
2.2 Microdissection and DNA extraction
Sequential sections (15µm-thick) from selected frozen blocks were prepared for microdissection of in situ and/or invasive lesions using a stereomicroscope to ensure tumor cell enrichment, as previously described (Sakr et al., 2014). The number of sections used for microdissection varied based on lesion size and cellularity with an average of 35 sections per lesion (range 6–89). DNA was extracted from microdissected samples of LCIS, ILC and normal breast ducts using the QIAamp DNA Micro Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Germline DNA was extracted from peripheral blood leukocytes using the QIAamp DNA Blood Maxi Kit (Qiagen). DNA was quantified using the Qubit Fluorometer (Life Technologies, Norwalk, CT).
2.3 Targeted capture massively parallel sequencing
Tumor and matched normal DNA obtained from 30 patients resulted in the availability of 85 samples (34 LCIS, 21 ILC, and 30 normal) for the purposes of this study (Table 1). Targeted capture massively parallel sequencing was performed on a platform containing baits for all exons of 273 genes recurrently mutated in breast cancer and involved in DNA repair pathways (Natrajan et al., 2014). Barcoded sequence libraries were prepared (New England Biolabs, Ipswich, MA; KapaBiosystems, Wilmington, MA) using at least 50ng of sheared DNA and pooled at equimolar concentrations into a single exon capture reaction (Roche NimbleGen, Madison, WI) (Cheng et al., 2015). Paired-end massively parallel sequencing was performed on a HiSeq2000 (Illumina, San Diego, CA).
Table 1.
Distribution of the 34 LCIS and 21 ILC samples obtained from 30 patients included in this study.
| Number of patients | Same quadrant | Different quadrant |
|
|---|---|---|---|
| LCIS only | 3 | - | - |
| ILC only | 6 | - | - |
| LCIS - LCIS pair | 6 | 1 | 5 |
| LCIS - ILC pair | 11 | 10 | 1 |
|
LCIS - LCIS - ILC triplet |
4* | - | 4 |
ILC, invasive lobular cancer; LCIS, lobular carcinoma in situ;
corresponding to 4 LCIS - LCIS pairs and 8 LCIS - ILC pairs.
Reads were aligned to the reference human genome GRCh37 using the Burrows-Wheeler Aligner (BWA) and sequencing data were processed by Genome Analysis Toolkit (GATK) (Li and Durbin, 2009; McKenna et al., 2010). Single nucleotide variants (SNVs) were defined using a combination of five mutation callers (i.e., MuTect, MutationSeq, GATK HaplotypeCaller, VarScan2 and SomaticSniper) (Cibulskis et al., 2013; Ding et al., 2012; Koboldt et al., 2012; Larson et al., 2012; McKenna et al., 2010). Small insertions and deletions (indels) were identified using the GATK HaplotypeCaller and VarScan2 (Koboldt et al., 2012; McKenna et al., 2010). Mutant allele fraction (MAF) was defined as the number of copies of the mutant allele divided by the number of copies of all alleles at a given locus. Mutations with allelic fraction of <5%, and/or those not detected by at least 2 of 5 mutation callers and not supported by at least 5 reads were disregarded. All candidate mutations were manually reviewed using the Integrative Genomics Viewer (Robinson et al., 2011). Copy number plots were generated using Varscan2, and were used for manual curation to determine whether a gene harboring a somatic mutation was also targeted by LOH.
Potential functional effect of each missense SNV was investigated using a combination of MutationTaster (Schwarz et al., 2010) and CHASM (Carter et al., 2009), and mutations identified as neutral/passengers by both computational prediction algorithms were considered passenger mutations, as previously described (Martelotto et al., 2014). The effect of in-frame indels was predicted using PROVEAN and MutationTaster (Choi et al., 2012; Schwarz et al., 2010). Genes affected by non-passenger mutations were further annotated using FATHMM (Shihab et al., 2013) and according to their presence in three cancer gene datasets: Cancer Gene Census (Futreal et al., 2004), 127 genes by Kandoth et al. (Kandoth et al., 2013) and the Cancer-5000S dataset by Lawrence et al. (Lawrence et al., 2014). “Lollipop plots” showing the distribution of mutations were generated using MutationMapper on cBioPortal (http://www.cbioportal.org/public-portal/mutation_mapper.jsp) (Gao et al., 2013).
2.4 Mutation validation
Samples with hotspot PIK3CA mutations and residual DNA were subjected to Sequenom MassARRAY (Sequenom) for validation of the PIK3CA mutations as previously described (Chandarlapaty et al., 2012; Sakr et al., 2014). Residual DNA from samples with CDH1 mutations were subjected to Sanger sequencing with primer sets (5’-CTGGGGTCCTCCCCAAT-3’ (forward), 5’-GGTGTGGGAGTGCAATTTCT-3’ (reverse)) as previously described (Weinreb et al., 2014). All analyses were performed in duplicate. Sequences of the forward and reverse strands were analysed using MacVector software (MacVector, Inc, Cary, NC) (Weinreb et al., 2014).
2.5 Clonality analysis
To infer clonal relatedness between lesions, we calculated for each mutation the probability of the mutation occurring in a pair of samples independently. Given that TCGA samples are unrelated, we chose to calculate each mutation’s rate of occurrence in the luminal-A subset of 235 samples (Cancer Genome Atlas, 2012): Mutation occurrence (MO) = number of mutations / 235 samples; Mutation’s rate of occurrence (MR) = MO × 100. The resulting probability of a pair to be independent (PI) based solely on that mutation would be the square of the mutation’s rate of occurrence in the mentioned subset: PI = MR × MR. Subsequently, we derived the probability of a pair to be clonal (PC) based on that mutation, by deducting the probability of a pair to be independent from 100: PC = 100 – PI. The total clonality confidence call is then the product of all shared mutations’ probability of clonality (PC): TPC = PC1 × PC2 × … × PCn (n = number of shared mutations). To further assess relatedness, we also performed unsupervised hierarchical clustering (Ward’s clustering algorithm with Euclidean distance) of samples harboring recurrent non-synonymous SNVs, silent SNVs and indels. We defined a recurrent SNV or indel as one that occurred in at least two samples.
2.6 Statistical analysis
Comparisons were assessed using Fisher’s exact and Chi-Square tests. 95% confidence intervals were adopted and p-values <0.05 were considered significant. Statistical analyses were performed using SAS software, version 9.1 (SAS Institute, Cary, NC).
3. RESULTS
Fresh-frozen tissue samples were prospectively harvested from 3 patients undergoing prophylactic mastectomy and 27 patients undergoing therapeutic mastectomy for ILC. From these 30 patients, we obtained DNA from 34 samples of LCIS and 21 samples of ILC (Table 1). This included 6 patients with paired LCIS samples (LCIS-LCIS), 11 patients with paired LCIS-ILC samples and 4 patients with triplets (2 LCIS and 1 ILC) resulting in a total of 19 LCIS-ILC comparisons and 10 LCIS-LCIS comparisons in the pairwise analysis. Selected samples from 4 cases reported here were also included in a parallel whole-exome sequencing analysis performed by our laboratory (Table 2, Table 3). The average size of the ILC lesions was 24 mm (range 14–75), and 26/26 (100%) of the ILC were estrogen receptor (ER)-positive/HER2-negative (Supplementary Table S1). ER and HER2 status were not assessed on the individual foci of LCIS. All LCIS and ILC lesions were E-cadherin negative by immunohistochemistry.
Table 2.
Shared mutations identified in pairs of LCIS - ILC lesions.
| Case number |
Lesion | Side Quadrant |
CDH1 | PIK3CA | CBFB | PKHD1L1 | RUNX1 | MAP3K1 | CDC25B | RELN | HERC2 | PCNXL2 | PDGFRA | ABCA13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | LCIS1¥ | R-UOQ | Q23* (21%) | H1047L (22%) | V4L (27%) | - | - | - | - | - | - | - | - | - |
| ILC¥ | R-UOQ | Q23* (34%) | H1047L (20%) | V4L (16%) | - | - | - | - | - | - | - | - | - | |
| LCIS2 | R-UIQ | Q23* (18%) | H1047L (22%) | V4L (19%) | - | - | - | - | - | - | - | - | - | |
| ILC | R-UOQ | Q23* (34%) | H1047L (20%) | V4L (16%) | - | - | - | - | - | - | - | - | - | |
| 13 | LCIS1 | L-UOQ | E336_splice (25%) | - | - | V1686I (17%) | - | - | - | - | - | - | - | - |
| ILC | L-UIQ | E336_splice (11%) | - | - | V1686I (16%) | - | - | - | - | - | - | - | - | |
| LCIS2 | L-LOQ | E336_splice (7%) | - | - | V1686I (10%) | - | - | - | - | - | - | - | - | |
| ILC | L-UIQ | E336_splice (11%) | - | - | V1686I (16%) | - | - | - | - | - | - | - | - | |
| 24 | LCIS1¥φ | R-LOQ | F730fs (17%) | H1047R (20%) | Q67H (26%) | - | - | - | - | - | - | - | - | - |
| ILC¥φ | R-LOQ | F730fs (10%) | H1047R (12%) | Q67H (20%) | - | - | - | - | - | - | - | - | - | |
| 28 | LCIS¥ | L-UIQ | D367fs (33%) | - | - | - | L71fs (29%) | - | - | - | - | - | - | - |
| ILC¥ | L-UIQ | D367fs (61%) | - | - | - | L71fs (36%) | - | - | - | - | - | - | - | |
| 29 | LCIS¥ | R-LIQ | Q23* (29%) | N345K (16%) | - | - | - | S416* (7%) | - | - | - | - | - | - |
| ILC¥ | R-LIQ | Q23* (48%) | N345K (28%) | - | - | - | S416* (16%) | - | - | - | - | - | - | |
| 31 | LCIS¥ | L-UOQ | E547* (35%) | - | - | - | - | - | R175Q (24%) | - | - | - | - | - |
| ILC¥ | L-UOQ | E547* (54%) | - | - | - | - | - | R175Q (33%) | - | - | - | - | - | |
| 33 | LCIS¥ | R-UOQ | - | - | - | - | - | - | - | - | - | R1215W (11%) | - | - |
| ILC¥ | R-UOQ | - | - | - | - | - | - | - | - | - | R1215W (14%) | - | - | |
| 38 | LCIS¥φ | R-UOQ | 522_splice (16%) | D350N (10%) E545K (15%) |
- | - | - | - | - | - | - | - | E289K (15%) | Q4805Q (14%) |
| ILC¥φ | R-UOQ | 522_splice (10%) | D350N (16%) E545K (20%) |
- | - | - | - | - | - | - | - | E289K (13%) | Q4805Q (9%) | |
| 48 | LCIS1¥φ | L-UOQ | 571_splice (56%) | - | - | - | - | - | - | - | - | - | - | - |
| ILC¥φ | L-UOQ | 571_splice (32%) | - | - | - | - | - | - | - | - | - | - | - | |
| 52 | LCIS¥ | R-UOQ | I248fs (21%) | H1047R (14%) | - | - | - | - | - | - | - | - | - | - |
| ILC¥ | R-UOQ | I248fs (22%) | H1047R (17%) | - | - | - | - | - | - | - | - | - | - | |
| 54 | LCIS¥ | L-UOQ | L714fs (6%) | E542K (13%) | - | - | - | - | - | - | R3668Q (14%) | - | - | - |
| ILC¥ | L-UOQ | L714fs (6%) | E542K (15%) | - | - | - | - | - | - | R3668Q (17%) | - | - | - | |
| 55 | LCIS¥ | R-UOQ | - | H1047R (19%) | - | - | - | - | - | T429S (20%) | - | - | - | - |
| ILC¥ | R-UOQ | - | H1047R (18%) | - | - | - | - | - | T429S (16%) | - | - | - | - |
ILC, invasive lobular cancer; L, left side; LCIS, lobular carcinoma in situ; LIQ, lower inner quadrant; LOQ, lower outer quadrant; MAF, mutant allele fraction; R, right side; UIQ, upper inner quadrant; UOQ, upper outer quadrant;
same quadrant;
samples also subjected to whole-exome sequencing. For each gene/ mutation, the amino acid change and the mutant allele fraction are shown. Shared mutations were identified in 14/19 pairs of LCIS-ILC.
Table 3.
─ Shared mutations identified in pairs of LCIS - LCIS lesions.
| Case number |
Lesion | Side-Quadrant | CDH1 | PIK3CA | CBFB | PKHD1L1 |
|---|---|---|---|---|---|---|
| 10 | LCIS-1 | R-UOQ | Q23* (21%) |
H1047L (22%) |
V4L (27%) |
- |
| LCIS-2 | R-UIQ | Q23* (18%) |
H1047L (22%) |
V4L (19%) |
- | |
| 13 | LCIS-1 | L-UOQ | E336_splice (25%) |
- | - | V1686I (17%) |
| LCIS-2 | L-LOQ | E336_splice (7%) |
- | - | V1686I (10%) |
|
| 53 | LCIS-1¥φ | R-UIQ | I178_splice (25%) |
- | - | - |
| LCIS-2¥φ | R-UIQ | I178_splice (7%) |
- | - | - |
ILC, invasive lobular cancer; L, left side; LCIS, lobular carcinoma in situ; LIQ, lower inner quadrant; LOQ, lower outer quadrant; MAF, mutant allele fraction; R, right side; UIQ, upper inner quadrant; UOQ, upper outer quadrant;
same quadrant;
samples also subjected to whole-exome sequencing. For each gene/ mutation, the amino acid change and the mutant allele fraction are shown. Shared mutations were identified in 3/10 pairs of LCIS-LCIS.
3.1 Targeted capture massively parallel sequencing analysis of LCIS
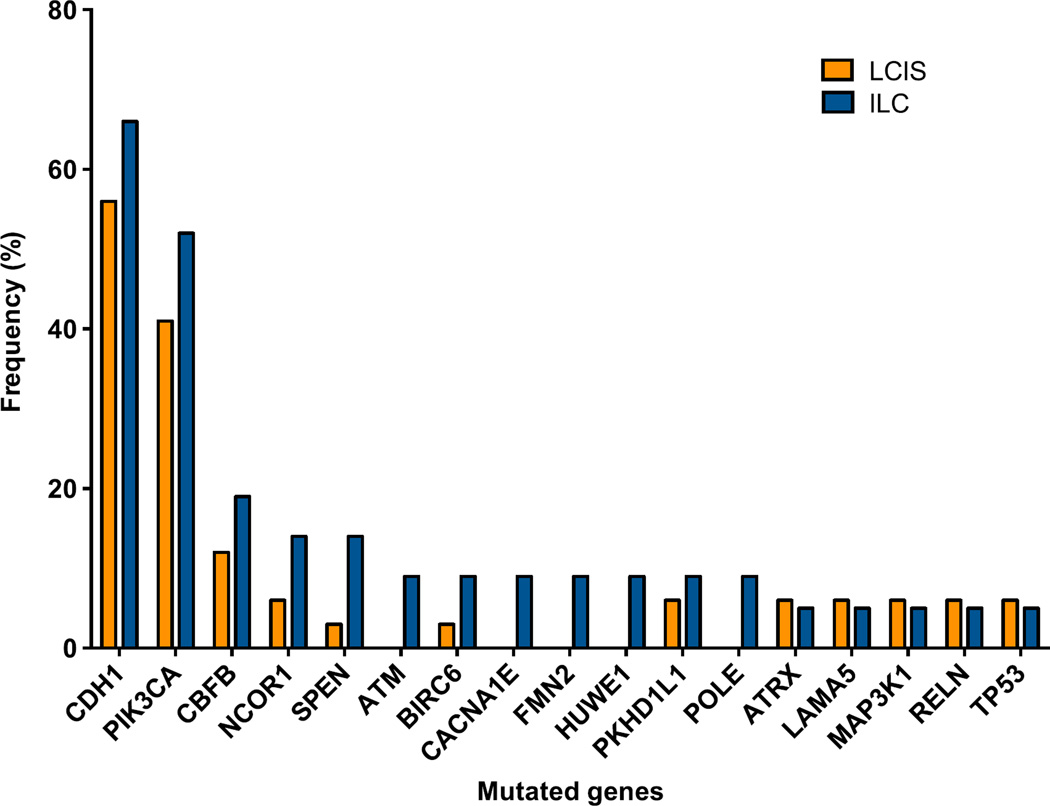
LCIS samples were sequenced to a median target depth of 238x (range 75x-603x), which resulted in the identification of 96 somatic mutations affecting 53 (19%) of the 273 genes analyzed. Of the LCIS samples analyzed, 82% (28/34) harbored at least one somatic mutation, and 68% (23/34) harbored two or more mutations. Of the 96 detected mutations, 66 (69%) were non-synonymous SNVs, 19 (20%) were indels and 11 (11%) were silent mutations. Genes recurrently affected by non-synonymous mutations in LCIS included CDH1, PIK3CA and CBFB with mutation frequencies of 56%, 41% and 12%, respectively (Figure 1, Supplementary Table S2). CDH1 gene mutations included 10 non-synonymous SNVs (10 splice-site) and 9 indels (8 frameshift, 1 splice-site) distributed among the multiple domains of the gene (Supplementary Figure S1); all CDH1 mutations were considered likely non-passenger events (Supplementary Table S3). LOH of 16q with CDH1 mutation was observed in 18/19 samples (Supplementary Table S3). PIK3CA gene mutations identified in LCIS included 15 non-synonymous SNVs (all missense) and 1 indel, of which 8 were found in the kinase domain (5 H1047R, 2 H1047L, 1 D1029H) and 5 in the helical domain (2 E542K, 1 E545K, 1 E545G, 1 Q546R); all mutations outside the helical and kinase domains (1 N345K, 1 D350N) and the indel (E110del) also outside the helical and kinase domains were considered to be non-passenger events (Supplementary Table S3); however, the biological impact of the indel identified remains to be determined. CBFB gene mutations included 3 non-synonymous SNVs (all missense) and 1 indel (splice-site), all considered to be non-passenger (Supplementary Table S3). LOH with CBFB mutation was observed in all 4 samples (Supplementary Table S3).
Figure 1. Mutational frequency of recurrently mutated genes in LCIS and ILC lesions.
Recurrently mutated genes identified by targeted massively parallel sequencing in 34 LCIS (orange) and 21 ILC (blue) analyzed in this study.
ILC, invasive lobular carcinoma; LCIS, lobular carcinoma in situ.
Among the 3 patients who underwent prophylactic mastectomy, there were 5 LCIS samples, of which one sample harbored a single mutation in the MAP3K1 (I1440T) gene and one sample displayed a mutation in CDH1 (H632fs), ERBB2 (L755S) and LAMA5 (P1241Q) (Supplementary Tables S3 and S4). All mutations except the LAMA5 mutation were considered to be likely non-passenger events (Supplementary Table S3).
PIK3CA mutations (H1047R, H1047L, E542K, D350N) in 8 LCIS samples with available residual DNA were confirmed by Sequenom analysis, and CDH1 mutation (Q23*) in 3 LCIS samples with available residual DNA was confirmed by Sanger sequencing analysis (Supplementary Table S3).
3.2 Targeted capture massively parallel sequencing analysis of ILC
ILC samples were sequenced to a median target depth of 307x (range 55x-665x), which resulted in the identification of 113 somatic mutations (80 non-synonymous SNVs, 12 indels and 21 silent mutations) involving 71 (26%) of the 273 genes analyzed. At least one non-synonymous mutation was identified in 19 (90%) samples and two or more non-synonymous mutations were found in 17 (81%) samples. As in LCIS samples, the genes recurrently affected by non-synonymous mutations were CDH1, PIK3CA and CBFB with mutation frequencies of 66%, 52% and 19%, respectively (Figure 1, Supplementary Table S2). CDH1 gene mutations included 8 non-synonymous SNVs (6 nonsense and 2 splice-site) and 6 indels (5 frameshift and 1 splice-site); all CDH1 mutations were considered likely non-passenger events (Supplementary Table S3). These 14 samples harboring CDH1 somatic mutations concurrently harbored LOH of the CDH1 wild-type allele. PIK3CA gene mutations included 11 non-synonymous SNVs (all missense) and 1 indel (E109del), with 4 mutations found in the kinase domain (3 H1047R, 1 H1047L) and 4 in the helical domain (1 E542K, 3 E545K). All PIK3CA mutations were considered non-passenger events (Supplementary Table S3). CBFB gene mutations included 4 non-synonymous SNVs, all missense and considered to be non-passenger events (Supplementary Table S3). LOH with CBFB mutation was observed in all 4 samples (Supplementary Table S3).
PIK3CA mutations (H1047R, H1047L, E545K, E542K, N345K) in 9 ILC samples with available residual DNA were confirmed by Sequenom analysis and CDH1 mutation (Q23*) in 2 ILC samples with available residual DNA was confirmed by Sanger sequencing analysis (Supplementary Table S3).
3.3 Comparison of the repertoire of somatic genetic alterations in LCIS and ILC
To define the differences in the repertoire of somatic genetic alterations found in LCIS and ILC, we first identified the genes commonly mutated between the LCIS and ILC analyzed. 30 genes were found to be affected by somatic mutations in both LCIS and ILC, including many genes reported to be recurrently mutated in ER-positive/luminal breast cancers (e.g., CDH1, ERBB3, GATA3, FOXA1, PIK3CA, MAP3K1, RUNX1, TP53, PTEN) (Banerji et al., 2012; Cancer Genome Atlas, 2012; Ellis et al., 2012; Stephens et al., 2012).
A comparison of the recurrently mutated genes in ILC and LCIS revealed no statistically significant difference between LCIS and ILC for any of the individual genes (Fisher’s exact test, p>0.5; Supplementary Table S2, Figure 1). Taken together, these findings demonstrate that LCIS and ILC harbor similar constellations of somatic mutations.
3.4 The majority of synchronous LCIS and ILC and independent ipsilateral foci of LCIS are clonally related
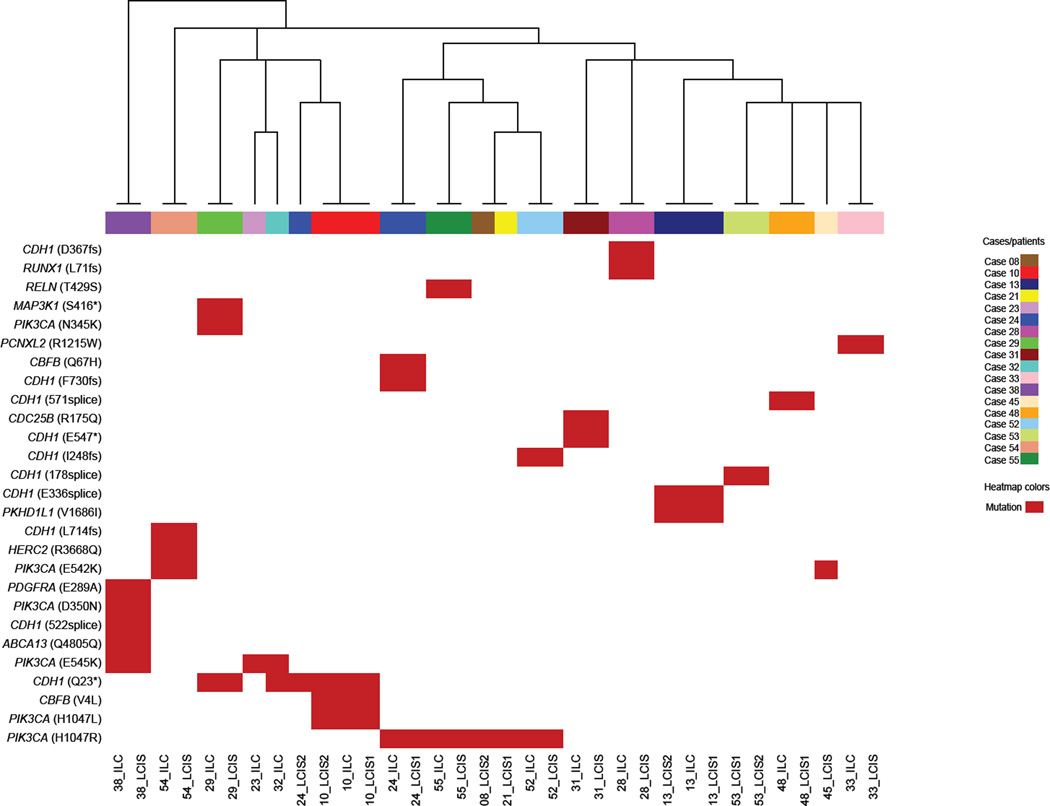
Pairwise analysis of the 19 LCIS-ILC paired lesions demonstrated at least one shared mutation in 14 (74%) sample pairs (Table 2). Among these 14 pairs, 7 LCIS-ILC pairs shared both identical CDH1 and PIK3CA mutations, 5 LCIS-ILC pairs shared a CDH1 mutation, and 1 LCIS-ILC pair shared a PIK3CA mutation. The analysis of the probability of a pair to be clonal based on number of shared mutations revealed that all 14 LCIS-ILC pairs were clonal, with a clonality confidence between 97% and 99.9% (Supplementary Table S5). Unsupervised hierarchical clustering of all LCIS and ILC samples included in this study using the most recurrently mutated genes (Figure 2) revealed that the 14 matched LCIS-ILC preferentially clustered together (13/14 pairs clustered together), providing additional evidence for the relatedness of these LCIS-ILC pairs. Anatomically, the paired lesions included in this study were located in the same quadrant of the breast in the majority of cases (11/14, 79% of LCIS-ILC pairs), and three pairs were located in different quadrants of the breast, the latter being derived from patients who underwent therapeutic mastectomy.
Figure 2. Unsupervised hierarchical clustering of LCIS and ILC samples using the recurrent somatic mutations identified by targeted massively parallel sequencing.
Hierarchical clustering of recurrent mutations identified in LCIS and ILC samples. Hierarchical cluster analysis was performed using Ward’s clustering algorithm with Euclidean distance. The colors in the phenobar represent lesions of a given case; the red box represents the presence of a mutation. Note that all paired LCIS-ILC samples cluster together.
ILC, invasive lobular carcinoma; LCIS, lobular carcinoma in situ.
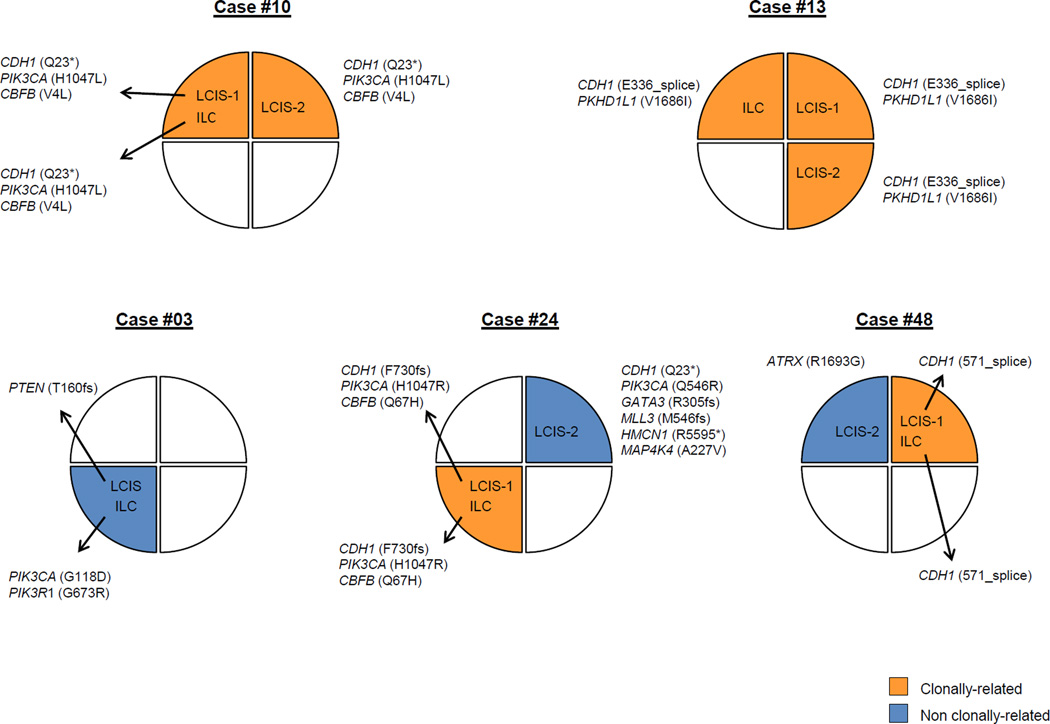
In three LCIS-ILC pairs, distinct mutations were observed in LCIS and ILC samples. In the first case (Case #03), the paired lesions were located in the same quadrant of the breast; however, the ILC harbored a PIK3CA and a PIK3R1 mutation, whereas the LCIS harbored a PTEN mutation (Figure 3). In the second case (Case #48), the lesions were located in different quadrants of the breast; whilst the ILC and LCIS-1 in the same quadrant harbored an identical CDH1 splice-site mutation, the LCIS-2 in a different quadrant harbored an ATRX mutation instead. In the third case (Case #24), we found that the ILC harbored a CDH1 (F730fs) and PIK3CA (H1047R) mutation distinct from those found in the LCIS-2 lesion in a different quadrant (CDH1 Q23*; PIK3CA Q546R). Furthermore, a CBFB mutation was found to be restricted to the ILC, while GATA3, HMNC1, MAP4K4, and MLL3 mutations were unique to the LCIS-2 lesion (Figure 3).
Figure 3. Clonal relatedness between LCIS and ILC.
Representation of the anatomical locations/breast quadrants of samples subjected to targeted massively parallel sequencing from cases for which three lesions (i.e., LCIS-LCIS-ILC) were analyzed or for cases where distinct somatic mutations between lesions were found. Clonally related lesions are indicated in orange, lesions in blue represent those without clonal relationship with any other lesion from the patient.
ILC, invasive lobular cancer; LCIS, lobular carcinoma in situ.
The remaining 2 LCIS-ILC pairs tested were equivocal with no mutations detected in one or both lesions of the pair (Supplementary Table S6).
We also compared multiple foci of LCIS from the same patient. Of the 10 LCIS-LCIS pairs included in this study, 3 (30%) pairs showed at least one common mutation (Table 3). The two LCIS lesions of the first pair (Case #53) were located in the same quadrant of the breast and shared an identical CDH1 I178 splice-site mutation. The second and third pairs were located in different quadrants of the breast and shared 2 (CDH1 and PKHD1L1) and 3 (CDH1, PIK3CA and CBFB) identical mutations, respectively (Table 3). The analysis of the probability of a LCIS-LCIS pair to be clonal based on the number of shared mutations revealed that all 3 cases the LCIS lesions were likely clonal with a clonality confidence between 98% and 99.9% (Supplementary Table S5). This was further supported by the observation that these 3 LCIS-LCIS pairs clustered together in the unsupervised hierarchical clustering performed (Figure 2 and Supplementary Figure S2). Conversely, distinct mutations were observed in 4 (40%) LCIS-LCIS pairs; in these pairs, the LCIS lesions were located in different quadrants of the breast. The remaining 3 (30%) LCIS-LCIS pairs were equivocal due to the lack of mutations in one or both of the lesions of the pair (Supplementary Table S6).
Overall, among all 17 LCIS-ILC or LCIS-LCIS pairs inferred to be clonal based on the number of shared mutations and unsupervised hierarchical clustering, 12 (71%) were located in the same quadrant of the breast, whereas 5 (29%) were found in different quadrants (Fisher’s exact test, 71% vs 29%, p=0.038). Of the 7 pairs determined to be non-clonal based on the presence of distinct mutations, 1 pair (case #03) was located in the same quadrant of the breast and 6 were in different quadrants of the breast (Fisher’s exact test, 16% vs 86%, p=0.029). There were no differences between clonal and non-clonal cases in terms of the clinical and histological features such as age, tumor size, ER/HER2 status and type of LCIS.
Finally, in 4 patients, we analyzed triplets of 2 LCIS and 1 ILC from the same breast. In case #10, all 3 lesions shared 3 common mutations (CDH1, PIK3CA and CBFB), and also in case #13 we identified 2 common mutations (CDH1 and PKHD1L1) in the 3 lesions (Figure 3). In case #48, however, LCIS-1 and ILC (located in the same upper outer quadrant) shared a common CDH1 mutation, which was not present in LCIS-2, located in the upper inner quadrant of the same breast (Figure 3). In case #24, different foci of LCIS harbored distinct mutations affecting the same genes (Figure 3); whilst LCIS-1 and the ILC, which were located in the right lower outer quadrant, shared the same somatic mutations in CDH1 (F730fs), PIK3CA (H1047R) and CBFB (Q67H); the LCIS-2, which was harvested from the upper inner quadrant of the same breast, harbored mutations in CDH1 (Q23*) and PIK3CA (Q546R) distinct from those found in the LCIS-1 and ILC, providing an example of a convergent phenotype in the development of LCIS.
4. DISCUSSION
Patients with LCIS have an increased risk of breast cancer that is 8- to 10-fold higher than the general population; yet the pathogenesis of this increased risk is poorly understood (Bratthauer and Tavassoli, 2002; Page et al., 1991). Morphologic and cytologic similarities between LCIS and ILC, combined with emerging reports of shared genomic alterations (mainly 1q gain, 16q loss and CDH1 mutations) between LCIS and synchronous ILC have reopened the debate about the precursor potential of LCIS (Dabbs et al., 2013; Hwang et al., 2004; Mastracci et al., 2006; Vos et al., 1997); however, most studies have been limited to a small number of cases and based on formalin-fixed paraffin-embedded samples. Here, using fresh-frozen tissue samples from 30 patients and a targeted massively sequencing approach, we provide evidence in favor of the hypothesis that LCIS is a non-obligate precursor lesion to invasive disease. In addition, we demonstrate that targeted capture massively parallel sequencing can be used to assess clonal relatedness between synchronous LCIS and ILC lesions. Taken together, our results demonstrate that LCIS is a non-invasive neoplastic lesion that displays a constellation of somatic mutations strikingly similar to that of ILC. Furthermore, our data provide evidence to suggest that LCIS is often clonally related to synchronous ILC, and anatomically, seemingly independent foci of LCIS harvested from the same breast may be clonally related.
In addition to the unique morphologic and cytologic description of the lobular phenotype, loss of E-cadherin protein expression has long been recognized as a hallmark feature of lobular disease (Dabbs et al., 2013; Foote and Stewart, 1941; Haagensen et al., 1978). Loss of E-cadherin expression is reported to occur through a combination of mechanisms including CDH1 gene mutation, allelic loss and CDH1 promoter methylation (Droufakou et al., 2001; Lopez-Garcia et al., 2010). Here we confirm that genomic alterations affecting CDH1 are an early event in lobular neoplasia with CDH1 gene mutations observed in 58% and 64% of LCIS and ILC lesions, respectively (Table 2, Figure 1). Further, there was no significant difference in the pattern of CDH1 mutations between LCIS and ILC samples. Previous studies with smaller sample sizes and different methodology have reported a higher frequency of CDH1 mutations in ILC (Logan et al., 2015); however, our findings are consistent with that of the most recent publication from The Cancer Genome Atlas Project, where CDH1 mutations were identified in 63% (80/127) of classic invasive lobular carcinomas (Ciriello G et al., 2014; Ciriello G, 2015).
Mutations in PIK3CA were the second-most frequent mutations in both LCIS and ILC with a mutation frequency of 39% and 55%, respectively (Table 2, Figure 1), which is also consistent with the frequency of PIK3CA mutations reported in ER-positive breast cancer and ILC specifically (Cancer Genome Atlas, 2012; Saal et al., 2005; Stemke-Hale et al., 2008). The CBFB gene, which, like CDH1, is located on chromosome 16, was also recurrently mutated in our sample set of LCIS and ILC with a mutational frequency of 11% and 23%, respectively (Table 2, Figure 1). The comparison of the mutational frequency of PIK3CA mutations in our dataset of ILC with that of luminal A ER-positive invasive carcinomas or the lobular TCGA dataset (Cancer Genome Atlas, 2012) revealed no significant difference. In contrast, mutations in CDH1 and CBFB were more frequent in ILCs in our dataset as compared to the luminal A ER-positive subtype from the TCGA dataset (p<0.0001). The increased prevalence of CDH1 mutations among lobular lesions is not unexpected and was confirmed in the TCGA lobular dataset (Ciriello G, 2015). The enrichment in mutations targeting CBFB in ILCs in our dataset, however, warrants further validation.
In a smaller study presented by our group, which included an independent cohort of patients with LCIS, CDH1 and PIK3CA were also found to be the most recurrently mutated genes in LCIS (De Brot et al., 2012). In this independent cohort, 36.4% (8/22) and 27.3% (6/22) of LCIS samples harbored PIK3CA and CDH1 mutations, respectively (De Brot et al., 2012), which is similar to the mutation frequencies identified in the current series (PIK3CA, Fisher’s exact test, two-tailed, p=0.785; CDH1, Fisher’s exact test, two-tailed, p=0.054).
Different methodologies have been used in the literature to assess clonal relatedness between LCIS and adjacent malignancies including ILC, yet most are based on the analysis of gene copy number alterations in formalin-fixed paraffin-embedded samples (Hwang et al., 2004; Morandi et al., 2006; Wagner et al., 2009), or on the presence of identical CDH1 mutations (Berx et al., 1996; De Leeuw et al., 1997; Droufakou et al., 2001; Rieger-Christ et al., 2001). One of the most frequently cited studies reporting the presence of shared mutations in LCIS and ILC included only 2 sets of paired lesions with common point mutations in the CDH1 gene (Vos, 1997 #1}. Nonetheless, given that CDH1 mutations in ILC do not occur at hotspots but at different residues within the gene, their specificity makes CDH1 highly informative when assessing clonal relatedness. In the current study, we used fresh-frozen samples of both LCIS and ILC lesions prospectively harvested from all quadrants of the breast and subjected them to targeted capture massively parallel sequencing. Common mutations observed in both lesions of LCIS-ILC pairs support a likely clonal relationship in 71% of cases, with paired lesions being in the same quadrant of the breast in 12/15 (80%) cases and the repertoire of shared mutations including either one or both of the most recurrently mutated genes CDH1 and PIK3CA (Table 3).
A limitation of our current study analysis was our inability to determine clonal relationship based on mutational data in 10% of LCIS-ILC pairs given the absence of mutations detected in one or both components of these pairs. Although targeted capture massively parallel sequencing allows a deeper sequencing coverage with lesser amounts of input DNA, it is limited to a panel of genes, and it is possible that our panel may not have included all of the genes relevant in the pathogenesis of LCIS or in the transition from LCIS to ILC. In addition, the small sample size reported here reflects the difficulty in harvesting fresh-frozen LCIS suitable for genomic analysis.
In conclusion, LCIS and ILC have a similar repertoire of somatic mutations, with PIK3CA and CDH1 being the most frequently mutated genes. The presence of identical mutations between LCIS-LCIS and LCIS-ILC pairs demonstrates that LCIS is a clonal neoplastic lesion and provides additional data that LCIS is a non-obligate precursor of ILC. Further, we demonstrate that targeted capture massively parallel sequencing can be used to assess clonal relatedness between paired lesions.
Supplementary Material
Highlights.
LCIS and invasive lobular carcinoma have a similar repertoire of somatic mutations
PIK3CA and CDH1 are the most frequently mutated genes in this setting
LCIS is a non-invasive clonal neoplastic lesion
LCIS may act as a non-obligate precursor of invasive lobular carcinoma
Acknowledgments
S. Piscuoglio is funded by a Susan G Komen Postdoctoral Fellowship Grant (PDF14298348). Research reported in this study was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- LCIS
lobular carcinoma in situ
- TCGA
The Cancer Genome Atlas
- ILC
invasive lobular carcinoma
- LOH
loss of heterozygosity, MSKCC, Memorial Sloan Kettering Cancer Center
- H&E
hematoxylin and eosin
- BWA
Burrows-Wheeler Aligner
- GATK
Genome Analysis Toolkit
- SNV
single nucleotide variant
- indels
insertions and deletions
- MAF
mutant allele fraction
- MO
mutation occurrence
- MR
mutation’s rate of occurrence
- PI
pair to be independent
- ER
estrogen receptor
Footnotes
The authors have no conflicts of interest to declare.
REFERENCES
- Andrade VP, Ostrovnaya I, Seshan VE, Morrogh M, Giri D, Olvera N, De Brot M, Morrow M, Begg CB, King TA. Clonal relatedness between lobular carcinoma in situ and synchronous malignant lesions. Breast cancer research : BCR. 2012;14:R103. doi: 10.1186/bcr3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortes ML, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Pina V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio RC, Parkin M, Pho NH, Quintanar-Jurado V, Ramos AH, Rebollar-Vega R, Rodriguez-Cuevas S, Romero-Cordoba SL, Schumacher SE, Stransky N, Thompson KM, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi DC, Richardson AL, Jimenez-Sanchez G, Lander ES, Gabriel SB, Garraway LA, Golub TR, Melendez-Zajgla J, Toker A, Getz G, Hidalgo-Miranda A, Meyerson M. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, Cornelisse C. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13:1919–1925. [PubMed] [Google Scholar]
- Bratthauer GL, Tavassoli FA. Lobular intraepithelial neoplasia: previously unexplored aspects assessed in 775 cases and their clinical implications. Virchows Archiv : an international journal of pathology. 2002;440:134–138. doi: 10.1007/s00428-001-0541-5. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer research. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sakr RA, Giri D, Patil S, Heguy A, Morrow M, Modi S, Norton L, Rosen N, Hudis C, King TA. Frequent mutational activation of the PI3K–AKT pathway in trastuzumab-resistant breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:6784–6791. doi: 10.1158/1078-0432.CCR-12-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O'Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PloS one. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Hoadley KA, Zhang H, Rhie SK, Bowlby R, Wilkerson MD, Kandoth C, McLellan M, Cherniack A, Laird PW, Sander C, King TA, Perou CM. Abstract S2-04: Comprehensive molecular characterization of invasive lobular breast tumors. Cancer research. 2014;75:S2–S04. [Google Scholar]
- Ciriello GGM, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GMK, Factor RE, Collins LC, Allison KH, Chen YY, Jensen K, Johnson NB, Oesterreich S, Mills GB, Cherniack AD, Robertson G, Benz C, Sander C, Laird PW, Perou CM. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbs DJ, Schnitt SJ, Geyer FC, Weigelt B, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Lakhani SR, Palacios J, Rakha E, Richardson AL, Schmitt FC, Tan PH, Tse GM, Vincent-Salomon A, Ellis IO, Badve S, Reis-Filho JS. Lobular neoplasia of the breast revisited with emphasis on the role of E-cadherin immunohistochemistry. The American journal of surgical pathology. 2013;37:e1–e11. doi: 10.1097/PAS.0b013e3182918a2b. [DOI] [PubMed] [Google Scholar]
- De Brot M, Andrade VP, Morrogh M, Berger MF, Won HH, Koslow MS, Qin L-X, Giri DD, Olvera N, Sakr RA, King TA. Cancer Res. San Antonio, TX: 2012. Novel mutations in lobular carcinoma in situ (LCIS) as uncovered by targeted parallel sequencing., Thirty-Fifth Annual CTRC-AACR San Antonio Breast Cancer Symposium. Abstract nr PD05-02. [Google Scholar]
- De Leeuw WJ, Berx G, Vos CB, Peterse JL, Van de Vijver MJ, Litvinov S, Van Roy F, Cornelisse CJ, Cleton-Jansen AM. Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. The Journal of pathology. 1997;183:404–411. doi: 10.1002/(SICI)1096-9896(199712)183:4<404::AID-PATH1148>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Ding J, Bashashati A, Roth A, Oloumi A, Tse K, Zeng T, Haffari G, Hirst M, Marra MA, Condon A, Aparicio S, Shah SP. Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinformatics. 2012;28:167–175. doi: 10.1093/bioinformatics/btr629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droufakou S, Deshmane V, Roylance R, Hanby A, Tomlinson I, Hart IR. Multiple ways of silencing E-cadherin gene expression in lobular carcinoma of the breast. International journal of cancer. Journal international du cancer. 2001;92:404–408. doi: 10.1002/ijc.1208. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote FW, Stewart FW. Lobular carcinoma in situ: A rare form of mammary cancer. The American journal of pathology. 1941;17:491–496. doi: 10.3322/canjclin.32.4.234. 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nature reviews. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagensen CD, Lane N, Lattes R, Bodian C. Lobular neoplasia (so-called lobular carcinoma in situ) of the breast. Cancer. 1978;42:737–769. doi: 10.1002/1097-0142(197808)42:2<737::aid-cncr2820420247>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Nyante SJ, Yi Chen Y, Moore D, DeVries S, Korkola JE, Esserman LJ, Waldman FM. Clonality of lobular carcinoma in situ and synchronous invasive lobular carcinoma. Cancer. 2004;100:2562–2572. doi: 10.1002/cncr.20273. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani SREIOSSJTPH, de van Vijver MJ, editors. IARC, Lyon: WHO Classification of Breast Tumors; 2012. [Google Scholar]
- Lakhani SR, Audretsch W, Cleton-Jensen AM, Cutuli B, Ellis I, Eusebi V, Greco M, Houslton RS, Kuhl CK, Kurtz J, Palacios J, Peterse H, Rochard F, Rutgers E, Eusoma The management of lobular carcinoma in situ (LCIS). Is LCIS the same as ductal carcinoma in situ (DCIS)? European journal of cancer. 2006;42:2205–2211. doi: 10.1016/j.ejca.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Lakhani SR, Collins N, Sloane JP, Stratton MR. Loss of heterozygosity in lobular carcinoma in situ of the breast. Clinical molecular pathology. 1995;48:M74–M78. doi: 10.1136/mp.48.2.m74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan GJ, Dabbs DJ, Lucas PC, Jankowitz RC, Brown DD, Clark BZ, Oesterreich S, McAuliffe PF. Molecular drivers of lobular carcinoma in situ. Breast cancer research : BCR. 2015;17:76. doi: 10.1186/s13058-015-0580-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchio C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–192. doi: 10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Ng C, De Filippo MR, Zhang Y, Piscuoglio S, Lim R, Shen R, Norton L, Reis-Filho JS, Weigelt B. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome biology. 2014;15:484. doi: 10.1186/s13059-014-0484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Shadeo A, Colby SM, Tuck AB, O'Malley FP, Bull SB, Lam WL, Andrulis IL. Genomic alterations in lobular neoplasia: a microarray comparative genomic hybridization signature for early neoplastic proliferationin the breast. Genes, chromosomes & cancer. 2006;45:1007–1017. doi: 10.1002/gcc.20368. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi L, Marucci G, Foschini MP, Cattani MG, Pession A, Riva C, Eusebi V. Genetic similarities and differences between lobular in situ neoplasia (LN) and invasive lobular carcinoma of the breast. Virchows Archiv : an international journal of pathology. 2006;449:14–23. doi: 10.1007/s00428-006-0192-7. [DOI] [PubMed] [Google Scholar]
- Natrajan R, Wilkerson PM, Marchio C, Piscuoglio S, Ng CK, Wai P, Lambros MB, Samartzis EP, Dedes KJ, Frankum J, Bajrami I, Kopec A, Mackay A, A'Hern R, Fenwick K, Kozarewa I, Hakas J, Mitsopoulos C, Hardisson D, Lord CJ, Kumar-Sinha C, Ashworth A, Weigelt B, Sapino A, Chinnaiyan AM, Maher CA, Reis-Filho JS. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. The Journal of pathology. 2014;232:553–565. doi: 10.1002/path.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DL, Kidd TE, Jr, Dupont WD, Simpson JF, Rogers LW. Lobular neoplasia of the breast: higher risk for subsequent invasive cancer predicted by more extensive disease. Human pathology. 1991;22:1232–1239. doi: 10.1016/0046-8177(91)90105-x. [DOI] [PubMed] [Google Scholar]
- Rieger-Christ KM, Pezza JA, Dugan JM, Braasch JW, Hughes KS, Summerhayes IC. Disparate E-cadherin mutations in LCIS and associated invasive breast carcinomas. Molecular pathology : MP. 2001;54:91–97. doi: 10.1136/mp.54.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer research. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- Sakr RA, Weigelt B, Chandarlapaty S, Andrade VP, Guerini-Rocco E, Giri D, Ng CK, Cowell CF, Rosen N, Reis-Filho JS, King TA. PI3K Pathway Activation in High-Grade Ductal Carcinoma In Situ--Implications for Progression to Invasive Breast Carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:2326–2337. doi: 10.1158/1078-0432.CCR-13-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human mutation. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung MC, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer research. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Oslo Breast Cancer C, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van 't Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos CB, Cleton-Jansen AM, Berx G, de Leeuw WJ, ter Haar NT, van Roy F, Cornelisse CJ, Peterse JL, van de Vijver MJ. E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. British journal of cancer. 1997;76:1131–1133. doi: 10.1038/bjc.1997.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner PL, Kitabayashi N, Chen YT, Shin SJ. Clonal relationship between closely approximated low-grade ductal and lobular lesions in the breast: a molecular study of 10 cases. American journal of clinical pathology. 2009;132:871–876. doi: 10.1309/AJCP7AK1VWFNMCSW. [DOI] [PubMed] [Google Scholar]
- Weinreb I, Piscuoglio S, Martelotto LG, Waggott D, Ng CK, Perez-Ordonez B, Harding NJ, Alfaro J, Chu KC, Viale A, Fusco N, da Cruz Paula A, Marchio C, Sakr RA, Lim R, Thompson LD, Chiosea SI, Seethala RR, Skalova A, Stelow EB, Fonseca I, Assaad A, How C, Wang J, de Borja R, Chan-Seng-Yue M, Howlett CJ, Nichols AC, Wen YH, Katabi N, Buchner N, Mullen L, Kislinger T, Wouters BG, Liu FF, Norton L, McPherson JD, Rubin BP, Clarke BA, Weigelt B, Boutros PC, Reis-Filho JS. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nature genetics. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.