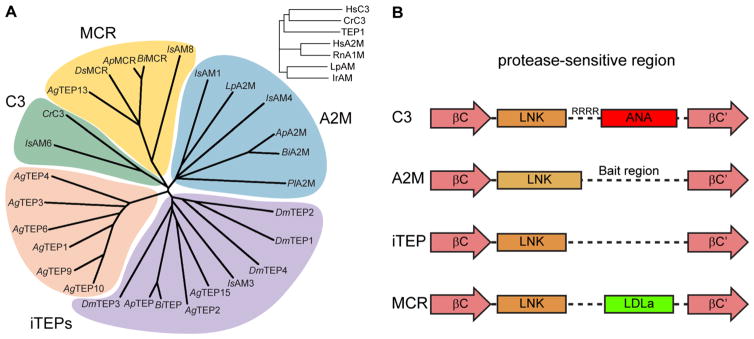
Fig. 3.
a Phylogenetic analysis of selected arthropod TEPs via Clustal Omega alignment of the MG3-ANK region. Crustacea: C. rotundica C3; L. polyphenus A2M; P. lacificas A2M. Arachnida: I. scapularis AM1, AM3, AM4, AM6, AM8 (Buresova et al. 2011). Hymenoptera: A. mellifera TEP (GB45417), A2M (GB42455), MCR (GB484204); B. impatiens TEP (BIMP24442), A2M (BIMP19175), MCR (BIMP19195). Diptera: D. melanogaster TEP1–4, MCR; A. gambiae TEP1–4, TEP6, TEP9–10, TEP12, MCR (TEP13), TEP14). Inset, A. gambiae TEP1 clusters with human and crustacean complement factors rather than A2Ms. b Schematic diagram of the protease-sensitive region for the four clades of TEPs found in arthropods. The LNK region of C3 and iTEPs are structurally homologous whereas the A2M LNK region is extended, terminating with a disulfide bond. A2Ms and iTEPs contain an unstructured region of broad protease-sensitivity while C3 and MCR have small, disulfide-rich domains between the LNK region and resumption of the MG6 fold