Fig. 4.
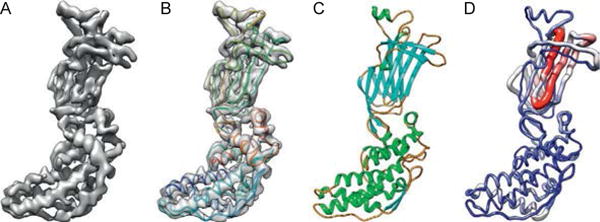
Modeling a 3.8 Å cryoEM map of VP6 of rotavirus. (A) A segmented density map of a capsid protein subunit of rotavirus (VP6) determined at 3.8 Å (EMDB1460). (B) A de novo model built by pathwalker superimposed on the density map. (C) A crystal structure of the same protein (PDB 1QHD). (D) A Cα rms deviation between the cryoEM model and crystal structure with the most and least deviation in red (gray in the print version) and blue (dark gray in the print version), respectively. These figures are kindly provided by Matthew L. Baker reproduced after Baker M.R. et al. (2012).
