Fig. 4.
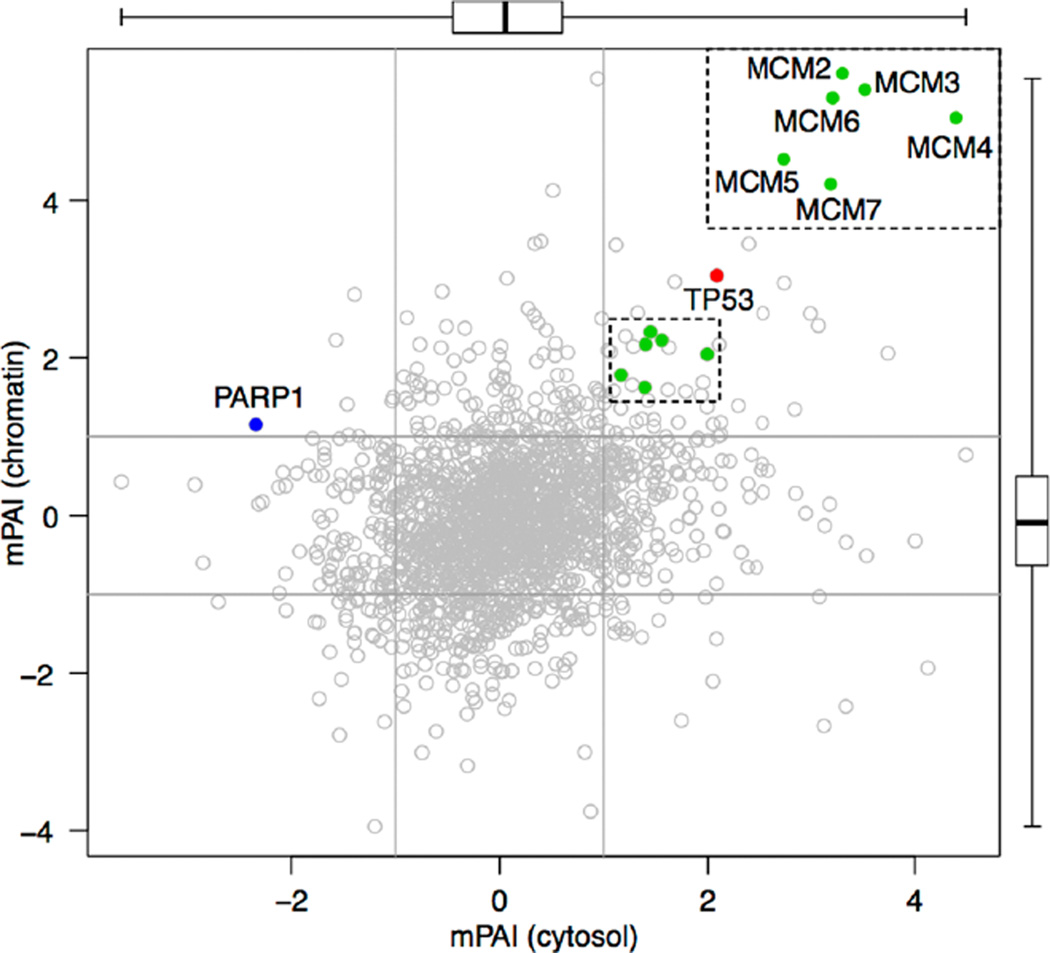
A double-positive mtp53 association seen for all MCM2–7 complex proteins in cytosol (x-axis) and nucleus (y-axis). Each dot (n = 1778) represents a protein with its position determined by its mPAI values in the cytosolic (x-axis) and the chromatin fractionations (y-axis). mPAI values were averaged if multiple peptides of the same protein were identified. Two side boxplots show the median, the lower and upper quartiles, and the range of mPAI values. The majority of points fall inside the x = −1, x = 1, y = −1, and y = 1 lines, indicating that abundance of most proteins are not significantly impacted by mtp53 knockdown in either fractionation. The mtp53 and the six members of the MCM2–7 complex fall in the top right quadrant, where protein levels are positively associated with mtp53 levels in both fractionations. PARP1 falls in the top left quadrant showing negative association with mtp53 in the cytosol but positive association in the nucleus, consistent with our previous experiment result.6 A searchable, zoomable, and clickable scatter plot of mPAI values for 4798 genes and 1330 associated pathways and gene sets is available at http://diverge.hunter.cuny.edu/~weigang/mpai-browser/