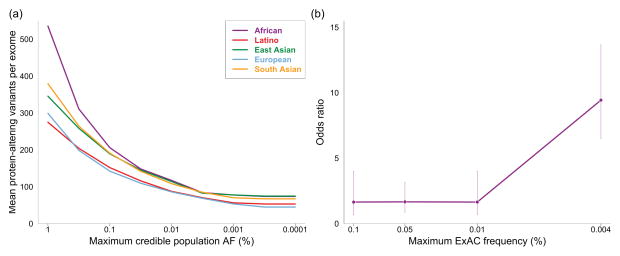
Figure 3.
The clinical utility of stringent allele frequency thresholds. (a) The number of predicted protein-altering variants (definition in Methods) per exome as a function of the allele frequency filter applied. A one-tailed 95% confidence interval is used, meaning that variants were removed from consideration if their AC would fall within the top 5% of the Poisson probability distribution for the user’s maximum credible AF (x axis). (b) The odds ratio for HCM disease-association against allele frequency. The prevalence of variants in sarcomeric HCM-associated genes (MYH7, MYBPC3, TNNT2, TNNI3, MYL2, MYL3, TPM1 and ACTC1, analysed collectively) in 322 HCM cases and 852 healthy controls were compared for a range of allele frequency bins, and an odds ratio computed (see Methods). Data for each bin is plotted at the upper allele frequency cutoff. Error bars represent 95% confidence intervals. The probability that a variant is pathogenic is much greater at very low allele frequencies.