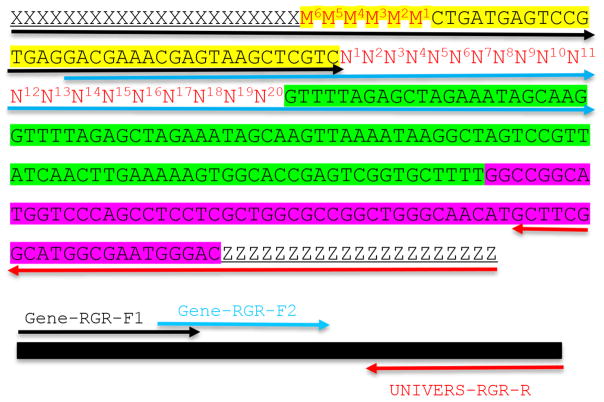
Figure 2. Molecular design of an RGR unit and the assembly of a complete RGR unit by PCR.
The top panel shows the complete sequence of an RGR unit that produces a gRNA that targets (N)20NGG sequence. The underlined sequences are adaptor sequences from the vector and are used for cloning the RGR unit into the final CRISPR vector. Note that the adaptor sequences vary accordingly if different vectors are chosen. N1 to N20 (in red) are the target sequence prior to the NGG PAM site. It is essential that the M6M5M4M3M2M1 and N1N2N3N4N5N6 are reverse complementary so that the ribozyme can undergoes self-cleavage as designed. Sequence highlighted in green is the core gRNA sequence. Sequence highlighted in yellow is the Hammerhead ribozyme. The HDV ribozyme is highlighted in purple. The primers are marked by different colors: black (Gene-RGR-F1), light blue (Gene-RGR-F2), and Red (UNIVERS-RGR-R). The Universal primer depends on the vector sequences and needs to be changed if different vectors are used. The complete RGR unit can be easily assembled by two over-lapping PCR reactions.