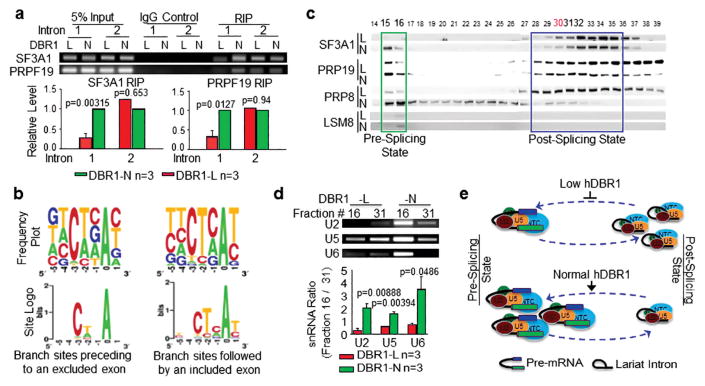
Figure 4. hDBR1 modulates the availability of splicing complexes in introns harboring a weak branch site.
(a) RNA immunoprecipitation (RIP) assays were performed using antibodies targeting SF3A1 or PRP19 in DBR1-L and N cells. The SF3A1 or PRP19 antibodies pulled down much greater amounts of pre-mRNA fragments composed of the 3′ end of FN-EDI intron 1 and the beginning portion of exon 2 in DBR1-N cells than in DBR1-L cells. RT-qPCR was used to detect the relevant RNA fragments. Top panels: representative qPCR products; Bottom panels: qPCR results from 3 separate experiments. The DBR1 RNA levels in the input samples and the transfection efficiency of the AS reporter are shown in Supplementary Figure S2a. (b) The RNA sequences of the branch sites in the introns followed by excluded or included exons showed weak or strong homology, respectively, to the authentic branch site. (c) Gel filtration analyses of DBR1-L and N cells for the dynamics of pre- or post-splicing complexes. (d) U2, U5, and U6 RNAs are accordingly altered in the two states of the splicing complexes. Top panels: representative qPCR products; Bottom panels: qPCR results from 3 separate experiments. (e) A schematic working model for a previously unknown function of DBR1: the level of DBR1 expression influences the dynamics of pre- or post-splicing complexes. When DBR1 expression levels are low, more snRNPs are retained in the post-splicing state, which affects the amount of snRNPs that are available to recycle back for the active splicing complex assembly.