Abstract
Intervertebral disc (IVD) degeneration is a significant contributor to low back pain. The IVD is a fibrocartilaginous joint that serves to transmit and dampen loads in the spine. The IVD consists of a proteoglycan-rich nucleus pulposus (NP) and a collagen-rich annulus fibrosis (AF) sandwiched by cartilaginous end-plates. Together with the adjacent vertebrae, the vertebrae-IVD structure forms a functional spine unit (FSU). These microstructures contain unique cell types as well as unique extracellular matrices. Whole organ culture of the FSU preserves the native extracellular matrix, cell differentiation phenotypes, and cellular-matrix interactions. Thus, organ culture techniques are particularly useful for investigating the complex biological mechanisms of the IVD. Here, we describe a high-throughput approach for culturing whole lumbar mouse FSUs that provides an ideal platform for studying disease mechanisms and therapies for the IVD. Furthermore, we describe several applications that utilize this organ culture method to conduct further studies including contrast-enhanced microCT imaging and three-dimensional high-resolution finite element modeling of the IVD.
Keywords: Bioengineering, Issue 122, intervertebral disc degeneration, organ culture, functional spine unit, mouse, lumbar, high-throughput
Introduction
Low back pain (LBP) is the leading factor for global disability and lost productivity in the workplace, and Americans alone spend in excess of 50 billion dollars on LBP treatment1. Although prevalent, the etiology of LBP remains complex and multifactorial. However, intervertebral disc (IVD) degeneration is among the most significant risk factors for LBP2.
The IVD is made of three microstructures: the exterior annulus fibrosis (AF), the interior nucleus pulposus (NP), and two cartilaginous endplates that sandwich the whole structure proximally and distally3. With aging and degeneration, the IVD components change structurally, compositionally, and mechanically4. These changes include the loss of proteoglycans and hydration in the NP, decreased disc height, and deteriorated mechanical competence5. These alterations are often accompanied by cytokines that promote an inflammatory response, as well as neutrophil and dorsal root ganglion intrusion into the joint space culminating in a cascade of events that lead to LBP symptoms6.
Studying the mechanisms of IVD degeneration is challenging in humans because it is often not possible to isolate the cause of the degeneration before the occurrence of low back pain. Thus, a reductionist approach of simplifying the experimental system down to the IVD organ allows mechanistic fine-tuning of causal variables and examining their downstream effects5. The system is reduced to only the native cell population and surrounding extracellular matrix, thus enabling the direct interpretation of the effects of external stimuli on IVD degeneration. Moreover, the lower cost and scalability of murine models, as well as the large number of genetically modified animals7, allow for the rapid targeted screening of IVD degenerative mechanisms and potential therapies. Here, we describe a murine organ culture system in which IVD cellular and tissue stability is maintained over 21 days, with specific focus given to the IVDs' homeostatic, mechanical, structural, and inflammatory patterns. Using this method, we monitor the IVDs' functional changes in a stab-induced injury model8 to understand the mechanisms behind disc degeneration. Furthermore, we describe several applications of this organ culture method to conduct further studies including contrast-enhanced microCT imaging and three-dimensional high-resolution modeling of the IVD.
Protocol
All animal experiments were performed in compliance with the Washington University in St. Louis Animal Studies Committee.
1. Animals
Obtain two strains of mice: 10-week old BALB/c (n = 6, BALB-M, BALB/cAnNTac) and 10-week old nuclear factor kappa-B-luciferase reporter animals (NF-κβ-luc) bred on a BALB/c background (n = 6, BALB/c-Tg(Rela-luc)31Xen).
Prior to dissection, euthanize animals with CO2 overdose at a flow rate of 2.5-3 L/min for 5 min followed by an additional 2 min of dwelling time.
Disinfect the outside region of the animal by bathing it in a 70% ethanol bath for 2 min before dissection.
2. Dissection
Make a longitudinal vertical cut on the dorsal surface of the mouse using small dissection scissors to expose the body cavity.
Make two longitudinal vertical cuts on either side of the animal's spine starting from the first lumbar vertebrae (L1) to the caudal vertebrae (C8).
Using a scalpel, fine forceps, and fine dissection scissors, carefully remove the spine from L1-C8 from the animal's body cavity.
Carefully remove excess soft tissues surrounding the spinal column, while ensuring to not scrape or injure the IVD on the ventral side.
Further dissect the spinal column into vertebrae-disc-vertebrae functional spinal units (FSUs) at the L1/L2, L3/L4, L5/L6, C3/C4, C5/C6, and C7/C8 discs.
Rinse the FSUs in Hank's balanced salt solution supplemented with 1% penicillin-streptomycin for 2 min.
Randomly assign the FSUs into three groups: uncultured and untreated FSUs (Fresh), cultured but untreated FSUs (Control), and cultured and stab treated FSUs (Stab).
Snap freeze the Fresh FSU samples with liquid nitrogen immediately after dissection and store in a -20 °C freezer.
3. Organ Culture Conditions
Prepare 500 mL of sterile culture media of 1:1 Dulbecco's modified Eagle's medium: Nutrient Mixture F-12 (DMEM:F12) supplemented with 20% fetal bovine serum (FBS) and 1% penicillin-streptomycin.
Using 10 mL serological pipettes, pipette 2 mL of culture media into each well of a 24-well culture plate.
Following dissection and rinse, place each FSU into an individual well in the media-filled 24-well plate.
Incubate the samples in a sterile incubator that maintains 37 °C, 5% CO2, 20% O2 and > 90% humidity.
After an initial culture period of 24 h, mechanically injure the Stab group samples via needle puncture of the annulus fibrosus using a sterile 27-gauge needle as shown in Figure 1A.
Change media every 48 h by preparing a new media-filled 24-well culture plate, and transfer samples from the old plate to the new using sterile tweezers. Aspirate the old media and dispose of the old culture plate in a biohazard waste bin.
On the final day of culture, incubate all samples in media containing 0.75 mg/mL nitro blue tetrazolium chloride for 24 h.
Afterwards, freeze all the FSUs with liquid nitrogen and store in a -20 °C freezer until ready for mechanical testing or histology (Figure 1B).
Figure 1. Experimental overview.
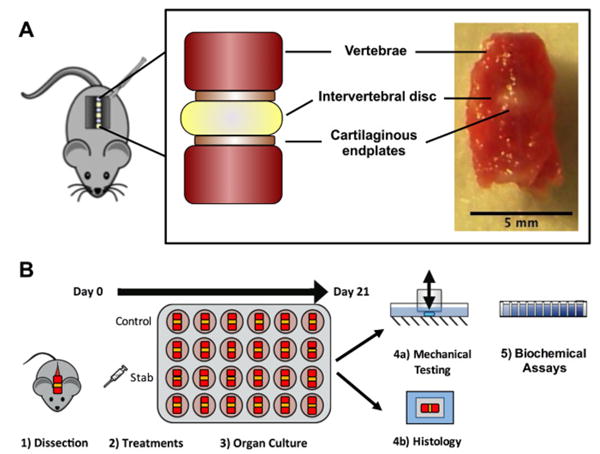
(A) Intervertebral disc functional spinal units (FSUs) were dissected from the lumbar and caudal segments; the FSUs contained two intact vertebrae, the cartilaginous endplates, and the intervertebral disc. (B) Following dissection, samples were divided into treatment groups and cultured in vitro for 21 days. Afterwards, a subset of samples was used for mechanical testing and biochemical assays, while another subset of samples was used for histological analysis.
4. Longitudinal Measurements NF-κB
Image the FSUs from NF-κβ-luciferase animals at 1, 5, 13, and 19 days to assess NF-κB expression.
Add 10 μL of 1 mg/mL luciferin solution to each well and incubate in a sterile incubator at 37 °C for 10 min.
Image the samples for bioluminescence using the imaging system with a 1 min exposure time and bin setting of 2.
Concurrently, use the machine to take a photograph and overlay it with the bioluminescence image to determine the anatomical location of luminescence in the IVD.
5. Mechanical Assessment
Determine the mechanical behavior of the IVDs by using displacement-controlled dynamic compression9.
Using a dissection microscope, scalpel, and tweezers, remove the bony vertebral bodies of the FSU from each sample while keeping the cartilaginous endplates intact and attached to the IVD.
Attach the isolated IVDs to a 1 cm × 1 cm × 0.2 cm aluminum plate using cyanoacrylate glue.
Measure the disc height and width by taking an average of three measurements on the longitudinal axis of each disc using a laser micrometer. Calculate the disc height ratio by dividing the average disc height by the maximum disc width.
Use the measured disc height to calculate the input strain values used in mechanical testing. Note that the average disc height was approximately 690 μm ± 39 μm.
Place the samples in a phosphate buffered saline bath under the compression machine and preload the disc to 0.02 N.
Cyclically compress the disc using a sinusoidal waveform at 1 Hz for 20 cycles at the 1% strain level and 5% strain level for 3 trials each and record the load and displacement values of the IVD. Allow 10 min of resting time between trials for the disc to relax and to prevent injury.
Calculate the average stiffness from the loading phase of the last cycle, and calculate the loss tangent from the phase angle between load and displacement data.
6. Proteoglycan and Collagen Quantification
Following mechanical testing, measure the disc wet weight by placing the isolated IVD in a pre-tared centrifuge tube and measuring the weight using an analytical balance.
Digest the isolated IVDs in 250 μL papain digestion solution (0.1 M sodium acetate, 0.01 M EDTA, 0.005 M cysteine HCl, pH 6.4) overnight at 65 °C using a block heater.
Centrifuge samples for 10 min at 2,000 × g and collect the supernatant fluid.
Measure the proteoglycan content of the IVD by using the dimethylmethylene blue (DMMB) assay with chondroitin sulfate standards.
Prepare DMMB solution (21 mg DMMB, 5 mL absolute ethanol, 2 g sodium formate in 1 L ddH2O).
Pipette 30 μL of standards and samples in triplicate in a 96-well plate, along with 250 μL of DMMB solution in each well.
Read the absorbance of the plate at 525 nm using a spectrophotometer.
Measure the collagen content using a hydroxyproline assay kit according to manufacturer instructions.
7. Histology
Fix a subset of samples in 4% paraformaldehyde for 24 h, de-calcify in 5% formic acid for 48 h, dehydrate with ethanol, and embed in paraffin.
Using a microtome, section the samples sagittally at 10 μm thickness and apply sections to glass slides. Stain the sections using Safranin-O/ Fast Green and DAPI.
Using a light microscope, image slides at 10× magnification.
8. Contrast-enhanced microComputed Tomography (microCT)
Scan a subset of samples at the 0, 2, 5, and 7-day time points using longitudinal Ioversol contrast-enhancement10, and terminal phosphomolybdic acid (PMA) contrast-enhancement at the 7 day time point.
4 h prior to microCT scan time, add 300 μL of 350 mg/mL iodine Ioversol solution to the culture media for a final concentration of approximately 50 mg/mL iodine-containing Ioversol solution and incubate in a sterile incubator at 37 °C for 4 h.
Following incubation, wrap the sample in sterile gauze and place in a 1 mL microcentrifuge tube.
Place samples in the microCT system and scan at 45 keVp, 177 μA, 10.5 μm voxel size, and 300 ms integration.
Export data from the microCT as DICOM files and visualize using software (e.g., OsiriX).
At the 7-day time point, fix samples using a 4% paraformaldehyde solution for 24 h, followed by 5% PMA solution for 3 days, and scan using the same settings as those used in step 8.4.
9. Three Dimensional Finite Element Modeling
Segment the DICOM files of the IVDs using software with manual thresholding (e.g., OsiriX).
Convert the NP and AF volumes of the IVD into voxel-based finite element meshes using Meshlab.
Combine the microstructures of the NP and AF to form a complete IVD and apply experimentally determined boundary conditions in the finite element software.
Representative Results
Figures 2-3 show representative results of proteoglycan distribution, NF-κB expression, stiffness, viscosity, disc height, and wet weight for cultured mouse IVDs. If cultured properly, the IVD parameters of the Control group should not be significantly different from the Fresh group. If the culture is infected or otherwise compromised, the Control group will be different from the Fresh group, especially in NF-κB expression and proteoglycan distribution (results not shown). Figures 4-5 show applications of organ culture to using contrast-enhanced microCT to obtain a three-dimensional model of the IVD; further, finite element modeling can be applied to these structures to determine tissue stress and strain distributions due to the global mechanical behavior.
Figure 2. Histology and NF-κB expression.
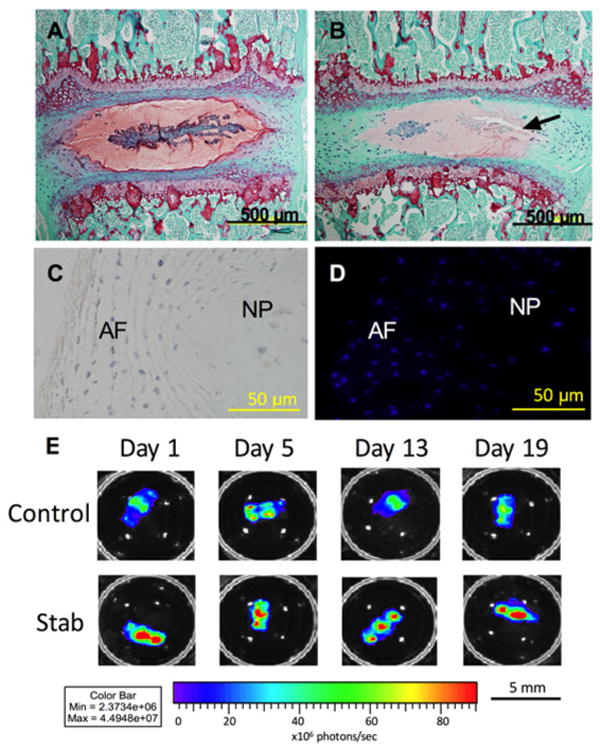
(A) After 21 days in culture, Safranin O staining (red) shows that proteoglycan content is maintained in both the AF and NP in the Control samples. (B) The Stab samples (puncture site indicated by arrow) showed decreased proteoglycan content in both the AF and NP. (C) Tetrazolium blue staining (blue) shows co-localization. (D) DAPI staining shows that cells are metabolically active and viable after 21 days in organ culture. (E) NF-κB expression in both the Control and Stab samples also indicate that the IVD is viable and responsive to its environment. The Stab samples have increased NF-κB expression at the 1, 5, 13, and 19-day time points. Lumbar IVDs are shown here.
Figure 3. Mechanics and composition.
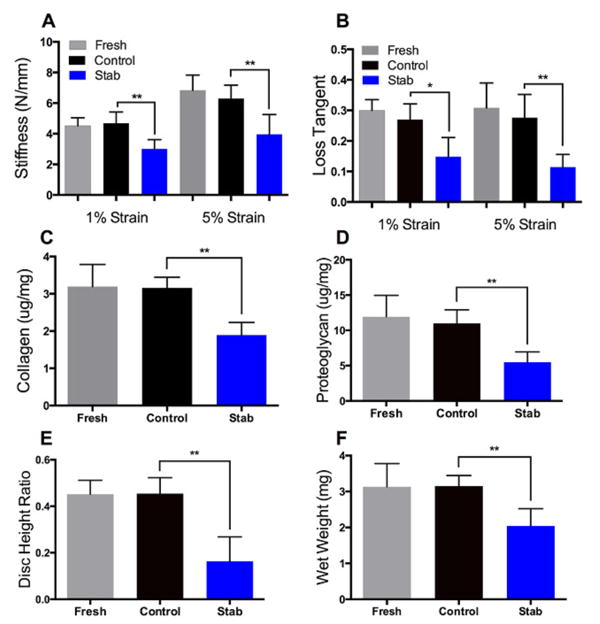
(A-B) Mechanical testing showed that at the 1% and 5% strain levels, Stab sample stiffness and loss tangent values are lower relative to Control samples. (C-D) Biochemical assays showed that compositionally, Stab samples had lower proteoglycan and collagen content (normalized to wet weight) relative to Control samples. (E-F) Structurally, Stab samples had a decreased disc height ratio and wet weight relative to Controls. Mechanically, compositionally, and structurally, Fresh and Control samples values were not significantly different from each other.
Figure 4. Contrast-enhanced microCT visualization.
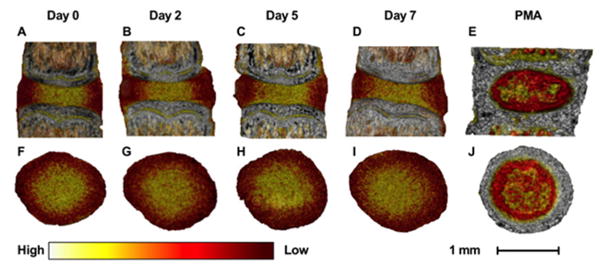
Contrast-enhanced microCT can be used to visualize the IVD in three-dimensions during the culture period. Caudal IVDs are shown here. The differential binding of Ioversol to the AF (lower attenuation) and NP (higher attenuation) allows one to distinguish the AF from the NP. Conversely, PMA preferentially binds to the collagen residues in the AF (higher attenuation), the endplates, and the notochord. The images were visualized using color such that white represents high attenuation, yellow represents medium attenuation, and red represents low attenuation. (A-E) Sagittal views of the IVD at days 0, 2, 5, and 7 with Ioversol contrast, and with PMA contrast. (F-J) Transverse views of the IVD at days 0, 2, 5, and 6 with Ioversol contrast, and with PMA contrast. Caudal IVDs are shown here.
Figure 5. Finite element modeling (FEM) of the IVD structure.
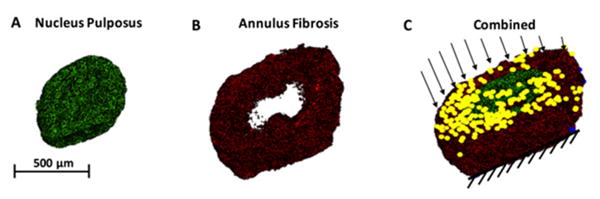
FEM analysis is a powerful mathematical tool that allows the computation of local material response to larger boundary conditions. For example, (A) the NP can be rendered separately from (B) the AF, and each compartment could be assigned unique constitutive properties. (C) A combined IVD structure with both the AF and NP is shown in 3D. When axial loads are applied to the disc on the superior endplate with the inferior endplate is a fixed edge, we can determine the local concentrations of stresses and strains. Yellow dots and black arrows represent a nodal load being applied.
Discussion
This protocol outlines an organ culture of the murine FSU with emphasis on monitoring the biological changes in the IVD. The successful maintenance of these cultures requires careful sterile techniques. In particular, the dissection steps 2.1-2.6 and the culture steps 3.1-3.6 require special care to ensure sterile conditions are maintained, and these steps should be performed preferably in an isolated procedure room with a HEPA airflow to minimize contaminants. Because the dissection process causes trauma to the tissues, the 24-h preconditioning period in 3.5 is required for all treatment groups including the controls. The viability assays can be used to ensure samples are alive and uncontaminated, and also serve as a confirmation that cellular homeostasis is maintained. The NF-κB luminescence readings can also be used to longitudinally monitor the inflammatory response of the cultured FSUs; an increase in NF-κB expression during culture could indicate the FSUs are contaminated or otherwise under stress from infection11, and we thus include it here as a possible troubleshooting step. Finally, comparisons to the Fresh groups serve as an additional check point for the Control cultured samples in histology and mechanical performance. This culture technique is amenable to multiple samples per plate, and in our experience, up to 24 samples on a 24-well plate can easily be accomplished. However, the multiplexing of samples also propagates the risk of cross-contamination and thus, it is recommended that FSUs be cultured on separate plates during troubleshooting. Additionally, while this protocol provides instructions for a culture period of 21 days, the same method can be used for culture periods that are longer or shorter. FSUs have been successfully cultured by our lab for culture periods ranging from 1 week to 5 weeks.
Like all models, organ culture models in murine have limitations, particularly in their ability to capture the loading conditions of the humans. There are also subtle differences in geometry between the murine and human IVDs12, and humans are bipedal with primarily axial loads on the spine while rats and mice are quadrupeds. However, since the cultured condition is relatively unloaded in its current form, the loading mode is likely to not have influenced the IVDs. Other systems such as bovine IVD organ culture models13,14 may be better suited for mechanical and loading interactions. Future work will incorporate loading conditions to enable the investigation of mechanobiological interactions between mechanical and environmental factors. The power of this approach lies in its consistency, versatility, and capability for high-resolution imaging. Our approach here demonstrates that it is possible to culture both lumbar and caudal IVDs, but it is important to note that these discs are anatomically different across aging and development15. As such, our experimental design randomizes the lumbar and caudal levels separately.
Contrast-enhanced microCT allow the high-resolution, nondestructive, three-dimensional imaging of the IVD. Different contrast agents can be used to highlight different compositional aspects of the IVD. We demonstrate here the use of Ioversol, an iodine-containing non-cytotoxic hydrophilic agent10, and phosphomolybdic acid (PMA), a heavy-metal molecule that chelates to collagen amino residues. Using these contrast-agents, the microstructural features in the IVD can be highlighted and identified. Ioversol differentiates the relative hydration of the IVD tissues, thereby providing contrast between the water-rich nucleus pulposus and the less hydrated annulus fibrosus. The PMA provides differentiation of relative collagen composition in tissues and will highlight the end-plates, annulus fibrosus, and the notochord. Ioversol can be used during culture to determine the longitudinal changes in hydration, while PMA can be applied to the IVD at the terminal point of culture to monitor collagen changes.
Future applications of this technology include utilizing other strains of transgenic and knockout mice to understand different aspects of IVD degeneration in other diseases. Further, it is a potential platform for co-culture with other organ systems and cells such as dorsal root ganglia to identify interactions that can contribute to IVD degeneration.
Supplementary Material
Acknowledgments
This work was supported by the Washington University Musculoskeletal Research Center (NIH P30 AR057235), Molecular Imaging Center (NIH P50 CA094056), Mechanobiology Training Grant (NIH 5T32EB018266), NIH R21AR069804, and NIH K01AR069116. The authors would like to thank Patrick Wong for his contributions in data collection.
Footnotes
Disclosures: The authors of this manuscript declare that they have no competing financial interests.
Video Link: The video component of this article can be found at https://www.jove.com/video/55437/
References
- 1.Dagenais S, Caro J, Haldeman S. A systematic review of low back pain cost of illness studies in the United States and internationally. Spine J. 2008;8(1):8–20. doi: 10.1016/j.spinee.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Urban J, Roberts S. Degeneration of the intervertbral disc. Arthritis Res Ther. 2003;5(6):1–48. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirza SK, White AA. Anatomy of intervertebral disc and pathophysiology of herniated disc disease. J Clin Laser Med Surg. 1995;13(3):131–42. doi: 10.1089/clm.1995.13.131. [DOI] [PubMed] [Google Scholar]
- 4.Acaroglu ER, et al. Degeneration and aging affect the tensile behavior of human lumbar anulus fibrosus. Spine. 1995;20(24):2690–2701. doi: 10.1097/00007632-199512150-00010. [DOI] [PubMed] [Google Scholar]
- 5.Abraham AC, Liu JW, Tang SY. Longitudinal changes in the structure and inflammatory response of the intervertebral disc due to stab injury in a murine organ culture model. J Orthop Res. 2016;34(8):1431–1438. doi: 10.1002/jor.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohtori S, Inoue G, Miyagi M, Takahashi K. Pathomechanisms of discogenic low back pain in humans and animal models. Spine J. 2015;15(6):1347–1355. doi: 10.1016/j.spinee.2013.07.490. [DOI] [PubMed] [Google Scholar]
- 7.Pelle DW, et al. Genetic and functional studies of the intervertebral disc: A novel murine intervertebral disc model. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, et al. Time course investigation of intervertebral disc degeneration produced by needle-stab injury of the rat caudal spine. J Neurosurg: Spine. 2011;15(4):404–413. doi: 10.3171/2011.5.SPINE10811. [DOI] [PubMed] [Google Scholar]
- 9.Liu JW, Abraham AC, Tang SY. The high-throughput phenotyping of the viscoelastic behavior of whole mouse intervertebral discs using a novel method of dynamic mechanical testing. J Biomech. 2015;48(10):2189–2194. doi: 10.1016/j.jbiomech.2015.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin KH, Wu Q, Leib DJ, Tang SY. A novel technique for the contrast-enhanced microCT imaging of murine intervertebral discs. J Mech Behav Biomed Mater. 2016;63 doi: 10.1016/j.jmbbm.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev: Immunol. 2009;9(11):778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell GD, Vresilovic EJ, Elliott DM. Comparison of animals used in disc research to human lumbar disc geometry. Spine. 2007;32(3):328–333. doi: 10.1097/01.brs.0000253961.40910.c1. [DOI] [PubMed] [Google Scholar]
- 13.Lee CR, et al. In vitro organ culture of the bovine intervertebral disc: effects of vertebral endplate and potential for mechanobiology studies. Spine. 2006;31:515–522. doi: 10.1097/01.brs.0000201302.59050.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan SC, Gantenbein-Ritter B. Preparation of Intact Bovine Tail Intervertebral Discs for Organ Culture. J Vis Exp. 2012;60:e3490. doi: 10.3791/3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holguin N, Aguilar R, Harland RA, Bomar BA, Silva MJ. The aging mouse partially models the aging human spine: lumbar and coccygeal disc height, composition, mechanical properties, and Wnt signaling in young and old mice. J Appl Physiol. 2014;116(12) doi: 10.1152/japplphysiol.01322.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


