Figure 4. NELF depletion leads to a genome-wide loss of transcription in progenitor cells.
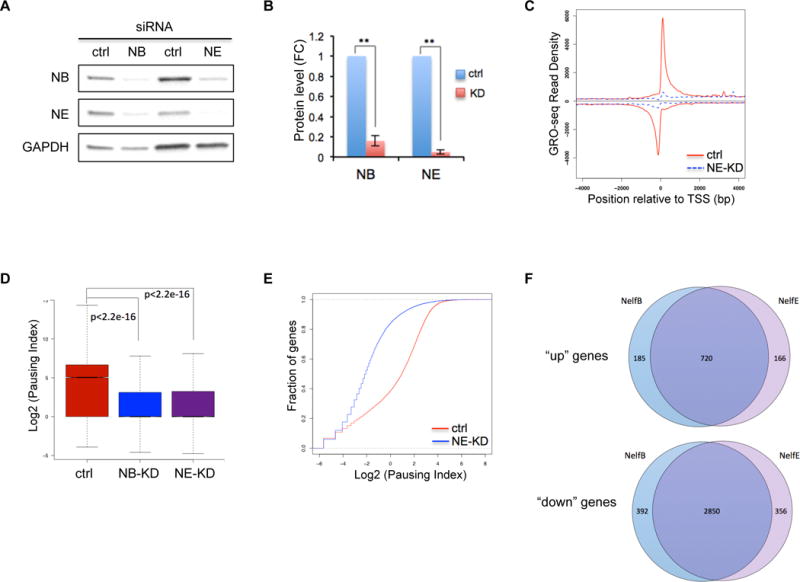
(A) Western blot of NELF-B and -E in human CD34+ HSPCs transfected with a scramble siRNA (ctrl), or siRNA targeting NELF-B or -E. Protein extraction was done on day 3 post-transfection. GAPDH is used as a loading control.
(B) Quantification of western blot in A by imageJ. Protein level is normalized to GAPDH and presented as fold change relative to control samples (n=3, mean ± SEM, **p<0.01).
(C) Metagene analysis showing Pol II occupancy measured by GRO-seq on both sense and anti-sense strands in human CD34+ HSPCs transfected with control-siRNA (ctrl) or siRNA targeting NELF-E (NE-KD). Cells were collected on day 3 post-transfection.
(D) Boxplot analysis to compare pausing index between cells transfected with a control-siRNA (ctrl), or siRNA targeting NELF-B (NB-KD) or –E (NE-KD).
(E) Cumulative distribution function analysis to compare PI distribution in cells transfected with a control-siRNA (ctrl), or siRNA targeting NELF-E (NE-KD)
(F) Venn Diagram showing the overlap of upregulated (“up”) and downregulated genes (“down”) between NELF-B and NELF-E knockdown cells.
