Abstract
Objective
We compared the efficiency of case selection strategies for following up a genome-wide linkage screen of multiplex families. We simulated datasets under three models by which continuous environmental or clinical covariates may contribute to disease risk or linkage heterogeneity: (i) a quantitative trait locus (QTL) underlying a continuous disease risk factor, (ii) a gene-environment interaction model, (iii) a heterogeneity model defined by distinct covariate distributions in linked and unlinked families.
Methods
Marker genotypes and covariate values were generated for affected sibling pair (ASP) families, according to the three models above. We evaluated two case selection strategies relative to a reference design, which compared all family probands to a sample of unrelated controls (‘all’). The first strategy ignored covariates and selected probands from families with NPL scores ≥ 0 (‘linked best’). The second strategy selected probands from families identified by an ordered subset analysis (OSA), which utilizes family-specific linkage and covariate information.
Results
The ‘linked best’ design provided power very similar to the ’all’ design under all three models. Under some QTL and heterogeneity models, the OSA design was both most powerful and most efficient.
Conclusions
Incorporating allele sharing and covariate information from ASP families into a case-control study design can increase power and reduce genotyping cost.
Keywords: Gene-environment interaction, Quantitative trait locus, Genetic heterogeneity, Linkage analysis, Ordered subset analysis, Study design, SIMLA
Introduction
Substantial resources have been invested in the collection of affected sibling pair (ASP) families to facilitate linkage analyses of complex human diseases. A whole-genome linkage analysis is often a first step toward the goal of identifying novel susceptibility genes, followed by association analysis. The linkage screen narrows the search by identifying genomic regions most likely to contain a disease susceptibility locus (DSL), and association analysis provides a much higher mapping resolution by virtue of linkage disequilibrium (LD) between alleles at genotyped marker(s) and the DSL. It is well established that case-control association analyses are more powerful than family-based association analyses in the absence of population stratification [1–3]. Recently, there has also been an interest in developing methods for the joint analysis of families, unrelated cases and unrelated controls [1, 4, 5]. In addition to the protection against population stratification provided by family-based association analysis [6], there is great practical value in continuing to work with the family datasets collected in the past for whole-genome linkage scans. This value is further enhanced by identifying study designs that make optimal use of the information about the likely DSL location provided by these family datasets.
Due to the complexity of the investigated phenotypes, it is important to incorporate environmental and clinical covariates into study design choices. This may include endophenotypes, if available. There are many different ways in which such covariates may either influence the disease risk directly, or partially explain the genetic heterogeneity commonly observed for complex diseases. The study presented here examined the efficiency of two case selection strategies under three plausible simulation models, referred to as ‘covariate models’, for genetically heterogeneous datasets: a quantitative trait locus (QTL) underlying a continuous covariate that is a risk factor for the disease, a multiplicative gene-environment interaction (GxE) model, and a heterogeneity model in which covariate distributions differ between linked and unlinked families, but in which the covariate does not influence the penetrance. The simulated datasets were analyzed by a two-stage approach, in which a stage 1 linkage analysis was followed by a stage 2 case-control association analysis that used one case per family. The two case selection strategies used only a subset of all family probands for association testing and were compared to a reference design, in which probands from all available families were compared to a sample of unrelated controls.
Materials and Methods
Data Simulation
With the simulation package SIMLA [7], we used a prospective logistic regression model as the penetrance function for binary disease outcomes generated on nuclear families with sibship size two. If D = 1 for affected and D = 0 for unaffected individuals, and the β parameters represent the natural logarithm of the odds ratios (ORs), this penetrance function can be written as
| (1) |
where x1 = 1 for the susceptible genotype(s), x1 = 0 for the referent genotype(s), and x2 is the value of a normally distributed continuous covariate E. As previously described, the x1 and x2 values for probands and non-proband pedigree members were assigned by appropriate simulation algorithms [7]. Covariate values were simulated so that 2.3% of the population were at the reference level (baseline risk) and 20% had a risk increase of at least OR(E) = exp(β2), compared to the baseline risk (see appendix for details). In SIMLA, this was accomplished by assigning the 80th percentile of the covariate distribution as the upper reference point [7]. Reference points, but not β2 values, were fixed across simulation models. For each replicate, 1,000 families with two affected siblings were retained for analysis to match standard linkage ascertainment strategies. Parents were assumed to be unavailable for genotyping.
Genotype and covariate data for 500 unrelated controls were generated conditional on their ‘unaffected’ status. We used a 50 cM map of 501 single-nucleotide polymorphism (SNP) markers spaced 0.1 cM apart, with all but one marker (SNP252) having two equally frequent alleles. The disease locus was located in the middle of the map at a distance of 0.05 cM from marker 252. The minor allele frequency (MAF) of SNP252 was chosen to be the same as the frequency of the disease susceptibility allele, and the extent of LD between marker and disease alleles was varied by specifying their founder haplotype frequencies in SIMLA according to r2 values of 0.05, 0.1 and 0.3. Disease genotypes were excluded from the analysis. All other marker loci were in linkage equilibrium with each other and with the disease locus, and Hardy-Weinberg equilibrium in the underlying population was assumed for all loci. We used the known (simulated) marker allele frequencies in the analysis files.
Simulation Models
Table 1 summarizes the three simulation models considered here. The models were chosen to yield a constant locus-specific sibling recurrence risk ratio, λs = 1.15, using the formulae in the appendix. Model 1 was a QTL model with three genotype-specific covariate means and standard deviations (SDs) for the general population. The covariate was assumed to be a risk factor for the disease with OR(E) > 1.0, and hence part of the penetrance function in equation (1), setting β1 = β3 = 0, β2 = ln(OR(E)) > 0. Thus, the QTL indirectly influenced the disease risk via the covariate effect on the penetrance, and the covariate (and QTL genotype) distributions differed between affected and unaffected individuals. The SD was assumed to be the same for the three QTL genotypes and determined the proportion of the variance explained by the linked QTL (heritability h2), with the total variance being a combination of major QTL and polygenic effects, nonspecific shared or unshared environmental factors and random error. Model 2 was a gene-environment interaction (GxE) model, in which the presence of GxE interaction between the DSL and continuous covariate was defined as more than multiplicative joint effects. As in the QTL model, the covariate was part of the penetrance function shown in equation (1) with β1 = β2 = 0, β3 = ln(OR(GxE)) > 0. In contrast, Model 3 was a heterogeneity model, in which linked (β1 = ln(OR(G)) > 0, β2 = β3 = 0) and unlinked families (β1 = β2 = β3 = 0 for the DSL linked to the marker map) were distinguished by covariate distributions with subgroup-specific means, with the same SD for each distribution. In this case, the covariate was not part of the penetrance function, and thus not a risk factor for the disease, and it did not have a genetic basis in the form of a QTL. Examples for such a covariate include age at onset, severity or clinical subtype (measured on a continuous scale) of the disease. We would like to point out that the size of the OR values in table 1 should be interpreted in conjunction with the assumed covariate distribution. The same λs value of 1.15 can be obtained with much smaller OR values by changing the reference points. For example, for Model 1 with α = 0.2, recessive inheritance, an overall λs = 1.15, corresponding to λs = 1.75 in the linked subset, can be obtained with OR(E) = 3 when 20%, instead of 2.3%, of the population are at the reference level (baseline risk) for the continuous covariate and 47.5%, instead of 20%, have at least a one-unit increase in risk. For Model 2 with α = 0.2, recessive inheritance, the same λs can be obtained with OR(GxE) = 3 with 20% of the population at baseline risk and 62.6% having at least a one-unit increase in risk.
Table 1.
Characteristics of simulated covariate models
| Model | α | Inheritance model | Allele frequency | OR(G) | OR(E) | OR(GxE) | QTL | SD (QTL) | h2 (linked families) | h2 (dataset) | µL (SD) | µU (SD) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.2 | dominant | 0.10 | 1.0 | 33.1 | 1.0 | yes | 4 | 0.49 | 0.10 | – | – |
| 0.2 | recessive | 0.35 | 1.0 | 38.5 | 1.0 | yes | 4 | 0.40 | 0.08 | – | – | |
| 0.5 | dominant | 0.05 | 1.0 | 8.6 | 1.0 | yes | 4 | 0.35 | 0.07 | – | – | |
| 0.5 | recessive | 0.35 | 1.0 | 10.5 | 1.0 | yes | 4 | 0.40 | 0.08 | – | – | |
| 2 | 0.2 | dominant | 0.10 | 1.0 | 1.0 | 18.2 | no | – | – | – | – | – |
| 0.2 | recessive | 0.35 | 1.0 | 1.0 | 25.8 | no | – | – | – | – | – | |
| 0.5 | dominant | 0.05 | 1.0 | 1.0 | 8.0 | no | – | – | – | – | – | |
| 0.5 | recessive | 0.35 | 1.0 | 1.0 | 10.0 | no | – | – | – | – | – | |
| 3 | 0.2 | dominant | 0.10 | 10.5 | 1.0 | 1.0 | no | – | – | – | 40 (5) | 20 (5) |
| 0.2 | recessive | 0.35 | 12.8 | 1.0 | 1.0 | no | – | – | – | 40 (5) | 20 (5) | |
| 0.5 | dominant | 0.05 | 5.2 | 1.0 | 1.0 | no | – | – | – | 40 (5) | 20 (5) | |
| 0.5 | recessive | 0.35 | 6.0 | 1.0 | 1.0 | no | – | – | – | 40 (5) | 20 (5) | |
Constants: disease prevalence 5%, locus-specific λs = 1.15. α: proportion of families linked to analyzed marker map. λs for the entire dataset was calculated as the weighted average of λs in the linked subset of families (1.75 for α = 0.2, 1.30 for α = 0.5) and λs = 1 in the unlinked subset. For Model 1, genotype-specific covariate means of 20, 30, and 30 were assumed for QTL genotypes aa, Aa, and AA in the dominant model, and means of 20, 20, and 30 were assumed in the recessive model, with the common standard deviation given in the SD(QTL) column. h2 is the theoretical heritability (additive and dominance variance component) that applies to the linked subset of families. h2 for the entire dataset was calculated as the weighted average of h2 in the linked subset of families and h2 = 0 in the unlinked subset. µL: covariate mean in linked families; µU: covariate mean in unlinked families. The ‘unlinked’ families (proportion 1-α), in which a second unlinked disease gene G2 was segregating, were generated with OR(G2) = 10, OR(E) = 1, RR(G2xE) = 1.
Data Analysis and Study Designs
For the stage 1 linkage analysis, we used the MERLIN software [8] to compute nonparametric multipoint lod scores derived from family-specific NPL scores, assuming the Sall scoring function under a linear model [9]. We included a subset of 50 SNPs (out of the 501 generated SNPs) evenly spaced 1 cM apart in the linkage map. The relationship between family-specific covariate averages and family-specific NPL scores was analyzed by OSA, using the high-to-low covariate ordering [10]. The OSA software reports the maximum nonparametric lod score for a covariate-defined subset of families and the map position at which it occurs. To obtain an empirical p value for a one-sided test of the OSA null hypothesis, which specifies no correlation of the family-level covariate with the family-specific evidence for linkage, a permutation test was employed. A minimum of 20 and maximum of 720 permutations were performed, which allows for the accurate estimation of p values on the order of 0.025 according to the precision criterion described previously [10]. We used the empirical p value as the criterion for significance, regardless of the size of the baseline lod score in the entire dataset or the size of the maximum lod score in the covariate-defined subset of families.
As the criterion for declaring the linkage analysis ‘successful’ and proceeding to case-control association analysis, we chose a lod score threshold of 1.0 for MERLIN, and a p value threshold of 0.05 for OSA. In replicates that met either of these criteria, the stage 2 case-control association analysis was performed on SNP markers located within a 10 cM region centered on the linkage peak. Three sets of cases and 500 unrelated controls were analyzed by logistic regression (SAS Institute, Cary, N.C., USA). Design A included probands from all ASP families, Design B included the single sibling classified as ‘linked best’ by MERLIN [11], which is equivalent to using probands from all families with non-negative NPL scores, and Design C included probands from the subset of families identified by OSA. The linkage peak was defined by the maximum lod score for all families in Designs A and B, and by the maximum lod score in the OSA-identified subset of families in Design C. We present detailed results from a logistic regression model that included a single SNP covariate with additive allele coding. Consistent with previous studies [12], we confirmed that this coding was most robust to deviation from the true model (dominant or recessive) used to generate the data (data not shown). For selected simulation models, we also generated results from a regression model that included two additional terms, a main effect term for the continuous covariate and a product term for SNP-covariate interaction.
Previous studies showed that the ascertainment (or analysis) of controls on the basis of their environmental covariates can improve the power to detect GxE interaction when main effects of genes and environmental factors have already been well established and interactive effects are the focus of the study [13, 14]. Therefore, we also evaluated the efficiency of control selection strategies for selected simulation models by performing separate association analyses of the three sets of cases defined above versus (i) controls from the lower 50% of the covariate distribution, and (ii) controls from the upper 50% of the covariate distribution.
Empirical Type I Error Rate, Power and Per-Genotype Information of Study Designs
It was previously shown that linkage and association test statistics are statistically independent under the null hypothesis of (i) no linkage and no association; (ii) linkage and no association; (iii) association and no linkage [15]. To verify these findings for our specific simulation models, we calculated the type I error rates of the three study designs by analyzing the non-associated markers in the 10 cM region around the linkage peak (all markers except SNP252) in 3,000 replicates. For these replicates, the association analysis was performed every time, not just in replicates that met the success criterion for the stage 1 linkage analysis. The empirical type I error rate was estimated as the proportion of replicates in which at least one non-associated marker in the 10 cM region centered on the maximum lod score on the chromosome met a Bonferroni-corrected p value threshold of 0.0005 (= 0.05/101) in the logistic regression analysis. Note that the stage 1 MERLIN and OSA analyses are tests of different null hypotheses, and that our goal was not to compare the power of these analyses for a more narrowly defined common null hypothesis. Instead, our goal was to examine how different case selection strategies influenced the probability of the stage 2 analysis to detect a true-positive association. Hence, the null hypothesis of interest for the different study designs is ‘no association’.
To estimate statistical power under the alternative hypothesis for the different simulation models of interest, 500 replicates were generated. Since the SNP with the smallest association p value in the region of interest is typically of greatest interest in practice, we defined the power of each study design as the proportion of replicates in which the associated SNP 252 met the design-specific linkage threshold, had the smallest case-control association p value of the 101 analyzed markers, and met a Bonferroni correction for multiple testing. When the linkage peak was located at a map position for which the 10 cM region centered on the peak did not exceed the end of the map on either side, we included in the association analysis the peak marker itself and 50 markers on either side, for a total of 101 markers and a Bonferroni-corrected threshold of 0.0005. When the linkage region did exceed the end of the map on either side, we still analyzed a total of 101 markers but the region was no longer symmetric around the peak.
The comparison of study designs in terms of absolute statistical power is only one aspect of practical interest. Another aspect is the per-genotype contribution to a case-control association test statistic, which is a measure of the relative power. For purposes of comparison with earlier work, we adopted a previously proposed measure for comparing case selection strategies [11], which is based on the following test statistic:
This statistic can be calculated from the design-specific estimated allele frequencies for the selected unrelated cases (one from each family) and controls, p̂case|design and p̂control, and the average number of cases Ncase|design (Ncontrol = 500 across all designs). Under Hardy-Weinberg equilibrium, as simulated here, this statistic is asymptotically equivalent to the Wald χ2 statistic for the SNP covariate in our logistic regression model. We verified empirically that the Wald χ2 statistic follows an asymptotic χ2 distribution on 1 d.f. at the non-disease-associated markers on our map (corresponding to the null hypothesis of equal allele frequencies in cases and controls) under the two case selection criteria employed in this study (data not shown). We did not use for formal hypothesis tests, but rather to calculate per-genotype contributions to this statistic as [11]. We then computed ratios of Idesign for Design B and Design C, relative to Design A (e.g., Rdesign B = Idesign B/Idesign A).
Results
The threshold of 1.0 for the nonparametric multipoint analysis was slightly liberal for a stand-alone linkage analysis, since it was exceeded in 8.2% of 10,000 replicates generated under the null hypothesis of no linkage for the particular marker map simulated here. The threshold of 0.05 for the OSA p value was previously shown to guarantee a type I error probability of 5% when only one covariate order is analyzed [10, 16]. In practice, an investigator would typically evaluate both high-to-low and low-to-high covariate orders and a 0.05 threshold, without adjusting for multiple testing, would then be slightly liberal. For the null hypothesis of ‘no association’ that is of greatest interest in this study, the empirical type I error rates for the three study designs ranged from 0.044 to 0.056, regardless of the linkage thresholds. Since both the MERLIN and OSA methods are based on linkage statistics, this was consistent with the previous report that linkage and association test statistics are independent under the null hypothesis of ‘linkage and no association’, and ‘no linkage and no association’ [15].
Figure 1–Figure 3 show the overall power of study designs A–C, estimated as the proportion of replicates in which the associated SNP 252 met the stage 1 linkage criteria, generated the smallest case-control association p value of all 101 analyzed markers, and met the Bonferroni correction for multiple testing. The two investigated proportions of linked families, α = 0.2 and α = 0.5, are shown on the x-axis, and the three sets of bars for each α value correspond to different levels of LD ranging from r2 = 0.05 to r2 = 0.3. The different bar types correspond to Designs A–C, i.e., case-control analysis comparing all family probands (Design A), only probands from families with non-negative NPL scores (Design B), and only probands from the subset of families identified by OSA (Design C) to the 500 unrelated controls.
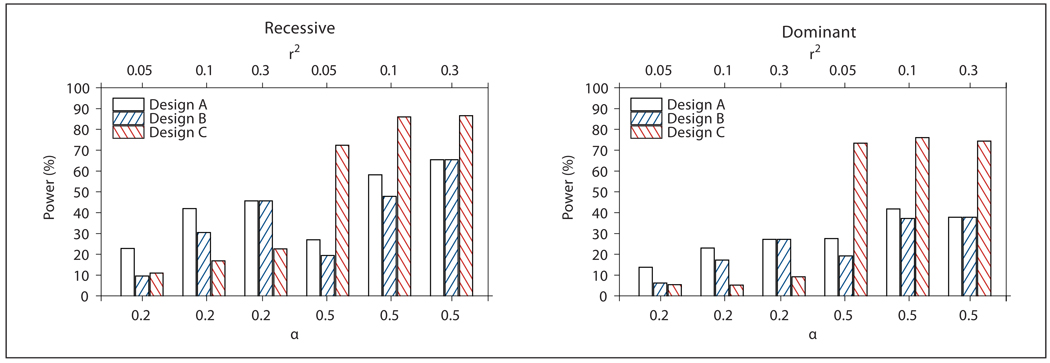
Fig. 1.
Power of Design A (all probands), B (probands from families with NPL ≥0) and C (probands from OSA-identified family subset) for Model 1 from table 1 (QTL). Power was defined as the proportion of replicates in which the associated SNP252 met the design-specific linkage threshold, had the smallest case-control association p value of the 101 markers analyzed by logistic regression, and met a Bonferroni correction for multiple testing. α: Proportion of linked families; r2: linkage disequilibrium between SNP252 and the QTL.
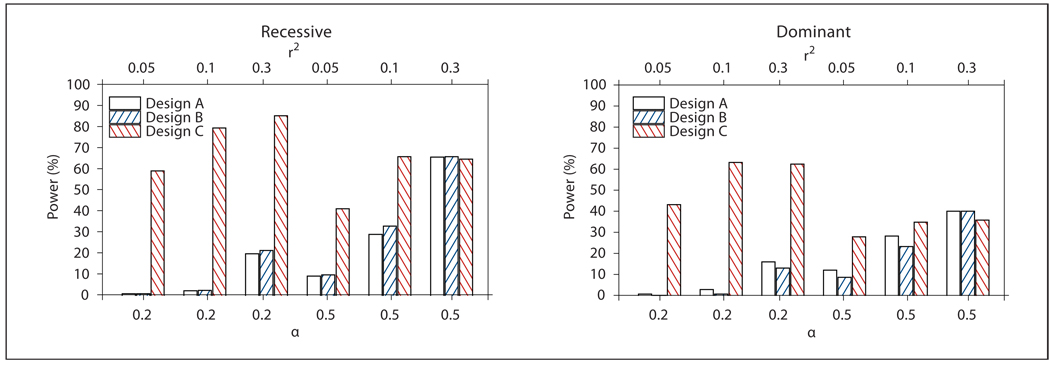
Fig. 3.
Power of Design A (all probands), B (probands from families with NPL ≥0) and C (probands from OSA-identified family subset) for Model 3 from table 1 (heterogeneity). Power was defined as the proportion of replicates in which the associated SNP252 met the design-specific linkage threshold, had the smallest case-control association p value of the 101 markers analyzed by logistic regression, and met a Bonferroni correction for multiple testing. α: Proportion of linked families; r2: linkage disequilibrium between SNP252 and the DSL.
Figure 1 shows the power of Designs A–C for the QTL simulation model (Model 1 in table 1), separately for the recessive and dominant inheritance model. For all designs, a nonparametric linkage analysis of the binary affection status had limited power to detect a QTL with the covariate model assumed here, especially with α = 0.2. Consistent with our previous findings [17], OSA had >70% power to detect linkage in a sample of ASP families when a trait determined by a QTL was analyzed as the OSA covariate and α values were on the order of 0.5. In this case, figure 1 shows that a selection of probands from OSA families (Design C) was substantially more powerful than an analysis of all probands, especially for low levels of LD. For example, for a recessive QTL model with α = 0.5, the power of Design C was ~72% for r2 = 0.05, compared to ~27% for Design A. The increase in power was especially remarkable considering that the average number of cases analyzed in Design C was much lower than in Design A, as illustrated in table 2. Across study designs and LD levels, Designs A and B had similar power, but Design B only analyzed 30% of the cases. It should be noted that the power of Design C depends on the chosen reference points for the continuous covariate distribution and the standard deviation (SD) within each genotype group, in addition to the choice of OR(E). For example, if at least 50% of the population had a one-unit increase in risk, instead of 20% as assumed here, a lower OR(E) generated equivalent power of OSA under the QTL model (data not shown). An increased SD for the genotype-specific distribution would decrease the calculated marginal OR(G), since the SD determines the reference points for the specified OR(E) (see appendix for details). To illustrate with an example, increasing the genotype-specific SD from 4 to 10 in Model 1 (α = 0.2, recessive), while holding all other parameters constant, changes the marginal OR(G) due to the QTL from 13.16 (table 2) to 6.22, using similar calculations as shown in the appendix.
Table 2.
Descriptive summary of three study designs
| Model | α | Genetic model | Design | p (true minor allele freq.) | p̂case | Avg. (SD) number of cases | p̂control (n = 500) | Avg. proportion of cases from linked subset | Rdesign | True marginal OR(G) |
ÔR(G) at SNP252 |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg. | SD | |||||||||||||
| 1 | 0.2 | dom | A | 0.1 | 0.173 | 1,000 (–) | 0.087 | 0.2 | 48.99 | 1 | 10.84 | 1.85 | 0.22 | |
| B | 0.1 | 0.181 | 307 (14.6) | 0.087 | 0.213 | 27.54 | 0.59 | 10.84 | 1.90 | 0.29 | ||||
| C | 0.1 | 0.268 | 248 (171) | 0.087 | 0.388 | 68.98 | 3.98 | 10.84 | 1.89 | 0.28 | ||||
| 0.2 | rec | A | 0.35 | 0.453 | 1,000 | 0.331 | 0.2 | 43.1 | 1 | 13.16 | 1.52 | 0.12 | ||
| B | 0.35 | 0.485 | 315 (14.4) | 0.331 | 0.239 | 38.38 | 1.46 | 13.16 | 1.56 | 0.18 | ||||
| C | 0.35 | 0.575 | 297 (196) | 0.331 | 0.349 | 94.08 | 8.97 | 13.16 | 1.69 | 0.15 | ||||
| 0.5 | dom | A | 0.05 | 0.168 | 1,000 (–) | 0.044 | 0.5 | 137.34 | 1 | 7.52 | 2.38 | 0.45 | ||
| B | 0.05 | 0.186 | 308 (13.7) | 0.044 | 0.514 | 70.05 | 0.48 | 7.52 | 2.55 | 0.58 | ||||
| C | 0.05 | 0.381 | 331 (204) | 0.044 | 0.713 | 285.12 | 7.78 | 7.52 | 3.79 | 0.85 | ||||
| 0.5 | rec | A | 0.35 | 0.534 | 1,000 (–) | 0.340 | 0.5 | 107.89 | 1 | 8.93 | 1.45 | 0.11 | ||
| B | 0.35 | 0.586 | 323 (14.8) | 0.340 | 0.538 | 100.87 | 1.59 | 8.93 | 1.54 | 0.15 | ||||
| C | 0.35 | 0.804 | 413 (211) | 0.340 | 0.705 | 518.56 | 37.95 | 8.93 | 1.85 | 0.19 | ||||
| 2 | 0.2 | dom | A | 0.1 | 0.172 | 1,000 (–) | 0.097 | 0.2 | 35.42 | 1 | 3.37 | 1.30 | 0.20 | |
| B | 0.1 | 0.182 | 307 (14.6) | 0.097 | 0.215 | 21.89 | 0.71 | 3.37 | 1.36 | 0.24 | ||||
| C | 0.1 | 0.249 | 297 (196) | 0.097 | 0.329 | 57.41 | 4.94 | 3.37 | 1.70 | 0.39 | ||||
| 0.2 | rec | A | 0.35 | 0.455 | 1,000 (–) | 0.346 | 0.2 | 33.92 | 1 | 4.44 | 1.16 | 0.09 | ||
| B | 0.35 | 0.486 | 315 (14.4) | 0.346 | 0.238 | 31.47 | 1.58 | 4.44 | 1.23 | 0.12 | ||||
| C | 0.35 | 0.545 | 297 (196) | 0.346 | 0.330 | 61.52 | 6.19 | 4.44 | 1.41 | 0.26 | ||||
| 0.5 | dom | A | 0.05 | 0.162 | 1,000 (–) | 0.047 | 0.5 | 117.38 | 1 | 3.32 | 1.91 | 0.35 | ||
| B | 0.05 | 0.183 | 308 (13.7) | 0.047 | 0.509 | 64.33 | 0.56 | 3.32 | 2.12 | 0.45 | ||||
| C | 0.05 | 0.231 | 331 (204) | 0.047 | 0.593 | 108.12 | 1.53 | 3.32 | 2.59 | 0.66 | ||||
| 0.5 | rec | A | 0.35 | 0.541 | 1,000 (–) | 0.344 | 0.5 | 110.94 | 1 | 4.03 | 1.31 | 0.10 | ||
| B | 0.35 | 0.594 | 323 (14.8) | 0.344 | 0.540 | 104.34 | 1.61 | 4.03 | 1.41 | 0.15 | ||||
| C | 0.35 | 0.630 | 413 (210) | 0.344 | 0.618 | 161.06 | 3.46 | 4.03 | 1.53 | 0.17 | ||||
| 3 | 0.2 | dom | A | 0.1 | 0.173 | 1,000 (–) | 0.087 | 0.2 | 48.99 | 1 | 2.90 | 1.32 | 0.18 | |
| B | 0.1 | 0.183 | 307 (14.9) | 0.087 | 0.219 | 28.54 | 0.63 | 2.90 | 1.36 | 0.23 | ||||
| C | 0.1 | 0.420 | 208 (81) | 0.087 | 0.822 | 166.75 | 24.54 | 2.90 | 2.65 | 0.57 | ||||
| 0.2 | rec | A | 0.35 | 0.455 | 1,000 (–) | 0.332 | 0.2 | 43.76 | 1 | 3.36 | 1.18 | 0.09 | ||
| B | 0.35 | 0.485 | 314 (14.3) | 0.332 | 0.239 | 37.79 | 1.37 | 3.36 | 1.24 | 0.13 | ||||
| C | 0.35 | 0.823 | 210 (65) | 0.332 | 0.916 | 423.98 | 198.37 | 3.36 | 1.94 | 0.28 | ||||
| 0.5 | dom | A | 0.05 | 0.166 | 1,000 (–) | 0.043 | 0.5 | 137.07 | 1 | 3.10 | 1.95 | 0.37 | ||
| B | 0.05 | 0.183 | 309 (13.9) | 0.043 | 0.512 | 69.24 | 0.47 | 3.10 | 2.12 | 0.47 | ||||
| C | 0.05 | 0.270 | 456 (145) | 0.043 | 0.912 | 200.29 | 3.35 | 3.10 | 2.88 | 0.60 | ||||
| 0.5 | rec | A | 0.35 | 0.538 | 1,000 (–) | 0.338 | 0.5 | 114.93 | 1 | 3.50 | 1.32 | 0.11 | ||
| B | 0.35 | 0.590 | 322 (14.2) | 0.338 | 0.543 | 105.95 | 1.55 | 3.50 | 1.43 | 0.15 | ||||
| C | 0.35 | 0.707 | 487 (102) | 0.338 | 0.948 | 311.98 | 11.2 | 3.50 | 1.65 | 0.17 | ||||
Average minor allele frequency at QTL or DSL in cases and controls, average number of cases, and average proportion of cases from linked subset of families are shown for each simulation model and study design (for r2 = 0.1). and Rdesign are defined in the Methods section. The true marginal OR(G) was calculated from the simulation parameters (see appendix for details), and the estimated ÔR(G) at SNP 252 (average and SD) was calculated from the simulated datasets. All averages were calculated across replicates that met the linkage thresholds for MERLIN (Designs A and B) or OSA (Design C), as explained in the text.
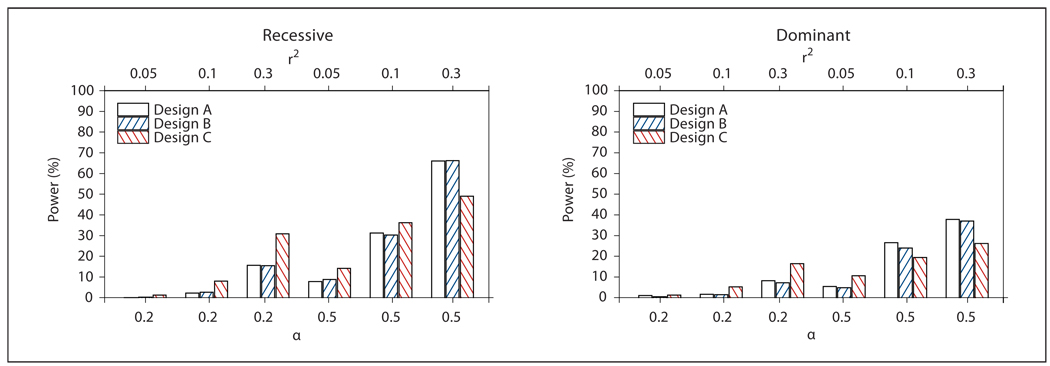
Figure 2 shows the power of Designs A–C for the GxE simulation model (Model 2 in table 1). Consistent with our previous findings for a ‘pure’ GxE interaction model with more than multiplicative joint effects in the absence of main effects [16], OSA did not substantially improve the power of a standard nonparametric analysis that ignores covariate values, regardless of LD levels. For α = 0.2, none of the designs had >31% power. For α = 0.5, r2 = 0.3 and a recessive model, Designs A and B achieved 66% power, the benefit of Design B again being an average ~30% reduction in the number of analyzed cases, while the power of Design C was 49%.
Fig. 2.
Power of Design A (all probands), B (probands from families with NPL ≥0) and C (probands from OSA-identified family subset) for Model 2 from table 1 (GxE). Power was defined as the proportion of replicates in which the associated SNP252 met the design-specific linkage threshold, had the smallest case-control association p value of the 101 markers analyzed by logistic regression, and met a Bonferroni correction for multiple testing. α: Proportion of linked families; r2: linkage disequilibrium between SNP252 and the DSL.
Figure 3 shows the power of Designs A–C for the heterogeneity simulation model, in which a main effect of the DSL linked to the marker map was only present in a proportion α of families (Model 3 in table 1). In this model, a remarkable power increase for Design C, compared to Designs A and B, was observed for α = 0.2. The power of Design C was 59–85% for the recessive and 43–63% for the dominant model across the three levels of LD, while the power of Designs A and B was <1% (recessive and dominant model) for very low LD (r2 = 0.05) and increased to only 15–20% for moderate LD (r2 = 0.3). Design C required genotyping an average of 200 cases for α = 0.2, compared to 1,000 cases for Design A and ~330 cases for Design B (table 2). The power increase for Design C was substantially reduced for α = 0.5, to the point where all three designs had very similar power (~65% for the recessive, 36–40% for the dominant model) with r2 = 0.3. The heterogeneity model clearly capitalizes on the strengths of the OSA method, especially when there is little overlap between linked and unlinked families and α is low [10], and it makes intuitive sense that the increased power of the OSA linkage analysis translates directly into increased power to detect association when only cases from the OSA-identified subset of families are compared to unrelated controls. As expected, the power of OSA decreased from ~80 to 70% when the standard deviation for the two covariate distributions in Model 3 (table 1) was increased from 5 to 10, holding α constant at 0.2 and r2 constant at 0.1.
Table 2 shows average estimated allele frequencies at the simulated QTL or DSL in the design-specific sample of analyzed cases and the 500 controls, and also compares the simulated ‘true’ marginal OR for the QTL or DSL to the average estimated OR for additively coded genotypes at SNP252. The biggest allele frequency difference in cases versus controls, and the highest proportion of cases from the linked subset of families, was obtained with Design C across all disease models. The variation in the analyzed number of cases was remarkably low for Design B for all models (SD across replicates on the order of 15), while the number of families (cases) identified by OSA varied substantially (SD across replicates on the order of 100–200) for all models except for Model 3 with α = 0.2. As expected, the estimated marginal ORs were much smaller for SNP252 than for the true QTL or DSL. The difference was especially pronounced for the QTL model (Model 1), for which the genotypes conferred an indirectly increased disease risk through the covariate (‘trait’) effect on the penetrance. The marginal OR in this model depends on the specified OR(E), the QTL allele frequency, the separation of genotype-specific means and the common SD of the respective normal distributions. The per-genotype contribution to the statistic for Design B and C, relative to Design A, was >1.0 for all recessive models, but <1.0 for Design B under the dominant model. This again illustrates the importance of the assumed allele frequencies, which were in the 0.05–0.10 range for the dominant models. In this case, the only slightly increased frequency of the ‘causal allele’ in the analyzed sample of cases was not sufficient to outweigh the increased variance due to the much smaller sample size (~300–330 cases in Design B, compared to 1,000 cases in Design A). Not surprisingly, by far the greatest ratio of the per-genotype contribution to the test statistic (~200) was observed for Design C under a recessive heterogeneity model with α = 0.2. However, the ratio for this OSA-based selection strategy was >1.0 even for the more challenging QTL and GxE models, with a range of 1.53 to 37.95 (table 2).
We examined whether the power of Designs B and C could be improved by selecting the affected sibling with the higher covariate value for the case-control association analysis. A small power gain on the order of 1 to 3 percentage points was observed for simulation Models 1 (QTL) and 2 (GxE). We also examined whether a selection of controls on the basis of covariate values was beneficial. To summarize our findings qualitatively, analyzing only controls from the lower 50% of the covariate distribution (for an average of 250 controls) provided very similar power as the analysis of all 500 controls for the QTL model. Analyzing only controls from the upper 50% of the covariate distribution (for an average of 250 controls) provided power very similar to the analysis of all 500 controls for the GxE model. Thus, the selection of controls on the basis of covariate values could in theory help further reduce genotyping costs, however, in the absence of knowledge about the true underlying model, it is preferable to genotype all available controls.
For the GxE model, we also investigated the power of the three designs to not only identify the disease-associated SNP (i.e., correct localization), but also to detect the presence of GxE interaction. For this purpose, the case-control data were analyzed with a logistic regression model that included a term for the continuous covariate and a product term for the covariate and the additively coded SNP genotype. This model provided very poor localization and power. The maximum proportion of replicates in which the p value at SNP252 for the estimate of either OR(G) or OR(GxE) was the smallest of all analyzed markers, and also smaller than 0.05, was only 14.2% across all models, heterogeneity parameters and study designs. Consistent with this observation, the Akaike Information Criterion (AIC) was always smallest for the most parsimonious analysis model that included only a term for the SNP genotype, compared to models with an additional term for the covariate, or two additional terms for the covariate and the SNP-covariate interaction.
Discussion
It has long been known that the analysis of cases from multiplex families enriches the sample for the presence of disease susceptibility alleles, leading to an increase in statistical power compared to the analysis of randomly sampled cases [2, 3]. We have extended this finding to show that the efficiency of case-control association study designs can be greatly improved when both allele sharing and covariate information are used to select cases from the same multiplex families that identified a linkage region for follow-up analysis. For all three investigated models by which clinical or environmental covariates may either influence the disease risk or capture linkage heterogeneity, selecting cases only from families with non-negative NPL scores provided very similar power as the analysis of all cases with only 33% of genotyped individuals. This is consistent with earlier reports for simulation models that did not include disease-associated covariates [11]. The OSA-based study design evaluated here selects cases not only on the basis of their IBD sharing with affected siblings, but also by evaluating the relationship between family-specific covariates and family-specific linkage evidence. Our results show that the selection of cases from the OSA-identified subset of families was beneficial when 40–60% of families were linked to a QTL for a continuous covariate. This covariate may have been measured in the family sample as a known risk factor for the disease of interest, or as an important endophenotype. In the presence of GxE interaction between a DSL locus and a measured continuous covariate, OSA was less helpful for case selection. This is consistent with our previous report that OSA has limited power to detect heterogeneity due to GxE interaction in a multiplicative penetrance model [16]. The greatest benefit of selecting cases from the OSA-identified subset of families was observed for a heterogeneity model with a small proportion (10–30%) of families in which a DSL linked to the marker map conferred a relatively strong main effect. In this model, linkage heterogeneity was captured by a measured covariate with distinct distributions in linked and unlinked families. The benefit of Design C was especially pronounced under a recessive inheritance model, presumably because ASPs are then more likely to share two alleles IBD, which provides more per-family information on linkage. The power of Design C is primarily driven by two factors: the difference in λs for the linked and unlinked families, i.e., 1.75 (for α = 0.2) or 1.3 (for α = 0.5) versus 1.0 in this study, and the extent of separation between the covariate distributions of these subsets. The number of multiplex families, and hence the number of cases in the OSA-identified subset of cases, is another important factor, and the formula for the statistic provides insight into the balance between increased susceptibility allele frequency in the subset versus increased variance due to a smaller sample size of cases. For both Design B and C, we found that case selection on the basis of their individual covariate values made little additional difference. This finding emphasizes the value of family-level information for enriching a sample of patients for inherited alleles.
A selection of controls on the basis of their individual covariate values allowed for a further increase in design efficiency, above and beyond that attributable to the case selection strategies. For the QTL model considered here, selecting the 50% of controls with the lowest covariate values increased the frequency of homozygous normal genotypes, and thus increased the MAF difference between cases and controls at the disease-associated SNP. For relatively common alleles, the increase in the MAF difference offset the increase in statistical variance due to a smaller sample size of controls, leading to virtually identical power estimates for analyzing all 500 controls versus only half of them. Conversely, for the GxE model, a selection of the 50% of controls with the highest covariate values increased the frequency of homozygous normal genotypes, with the same effect of an increased MAF difference between cases and controls. However, relative to the QTL model, the absolute MAF difference was smaller for the GxE model. These results suggest that control selection on the basis of covariate values may in principle allow for additional savings in genotyping costs, but in the absence of knowledge about the true covariate model (e.g., QTL vs. GxE), it is preferable to genotype all available controls. At the statistical analysis stage, an analysis of all controls compared to an analysis of controls from the lower or upper 50% of the covariate distribution may provide insight into the possible nature of the underlying covariate model. It may also be helpful to visually compare the relationship between marker genotypes and covariate values in affected and unaffected individuals with our recently developed software tool SIMLAPLOT [18].
The main limitation of our simulation study was the assumption of a very simple marker spacing and LD structure for the genomic region of interest. Only a single SNP was in LD with the minor allele at the QTL or DSL, while all other SNPs were in linkage equilibrium with each other and with the causal allele. This made it possible to detect association by analyzing one SNP at a time, and we did not consider the additional challenges posed by situations in which only a haplotype of several SNPs may be in LD with an unknown susceptibility variant. In this simple situation, the Bonferroni correction is not overly conservative. However, for the complex LD structure of the human genome, more sophisticated multiple testing corrections are desirable in order to balance statistical power and false-positive rate, and this continues to be an active area of methodological research motivated by the current interest in whole-genome association studies.
Our findings have several implications for applied studies of complex human diseases. First, they emphasize the value of low-density whole-genome linkage screens with much lower per-sample cost, even at a time when whole-genome association screens have become technically feasible. The analysis of cases selected on the basis of linkage evidence can provide substantial power increases for localizing a QTL or DSL with high resolution, even at low levels of LD and in the presence of substantial genetic heterogeneity. Our findings should extend from ASP datasets typical of late-onset disorders (i.e., without genotyped parents, as simulated here) to those typical of early-onset disorders, since the availability of parental genotypes improves the power of linkage analysis.
Second, our results suggest that investigators may want to either consider the construction of region-specific sample lists for follow-up genotyping, or, when that is impractical in terms of sample or project management, to use linkage results as a guide for the inclusion or exclusion of cases at the statistical analysis stage.
Third, our study illustrates the difficulty of detecting GxE interaction with the same dataset used for gene discovery. A logistic regression of cases and controls that included only a single SNP genotype term in the model was much more powerful in terms of gene localization than a model that included additional covariate and interaction (product) terms. This suggests that gene discovery and a more detailed modeling of identified candidate genes, including estimation of penetrance, attributable risk, and interaction with environmental factors, are best performed in independent large datasets. This, in turn, emphasizes the importance of ascertaining unrelated cases, with or without sampled relatives, and appropriately matched unrelated controls in parallel with multiplex families for linkage analysis [19]. From a practical perspective, we note that multiplex families for a genome-wide linkage analysis are typically more difficult to collect than unrelated cases. A smaller sample size of multiplex families limits the power of the study design we have evaluated since it is well known that the detection of GxE interaction requires substantially larger sample sizes than the detection of main effects, particularly when measured SNPs are not in perfect LD with the true DSL. It would be beneficial to evaluate in future studies under which conditions the OSA-identified covariate cutoff point is useful to select a subset of all available cases, including those without affected sampled relatives, in order to decrease the genetic heterogeneity of the case sample. Extensions of the OSA method to family-based or case-control association mapping are also desirable.
Finally, our findings emphasize the importance of collecting environmental and clinical covariate data in gene discovery studies, in addition to the primary diagnostic criteria for determining affection status. The incorporation of family-level covariate information appears to contribute more strongly than individual covariate levels to the enrichment of a sample of patients for inherited alleles. The importance of obtaining detailed phenotypic data for studies of complex human diseases with substantial genetic heterogeneity cannot be overestimated. Covariate information can also be extremely useful in terms of illuminating biological mechanisms or pathways that may be important contributors to the disease risk in a subset of patients and families. For example, a spectrum of sequence variations in the proprotein convertase subtilisin/kexin type 9 serine protease gene (PCSK9) with a wide range of allele frequencies and effect sizes was shown to contribute to inter-individual differences in low-density lipoprotein cholesterol (LDL-C) levels [20]. Variants associated with a reduction in mean LDL-C were indirectly associated with a reduction in the risk of coronary heart disease ranging from 47% for white subjects to 88% in black subjects [21]. Studying the variation in a continuous disease risk factor in unselected samples allows for a more comprehensive assessment of genotype-phenotype relationships. However, consistent with the findings reported here, a judicious selection of cases and controls typically provides a much more efficient study design for gene identification since most of the information and statistical power is provided by individuals in the tails of the distribution [22].
Of relevance to this study, novel statistical methodology has recently been developed to simultaneously test for linkage and LD in datasets that include variable pedigree structures (affected sibling pair families, singleton families with case-parent triads and/or discordant sibling pairs) as well as unrelated cases and controls [1]. If it is financially feasible to genotype such datasets with SNP panels of sufficiently high density to detect association signals due to LD with untyped susceptibility loci, this methodology is very promising and appears to be statistically powerful [23]. In the presence of financial constraints, however, we believe that the two-stage linkage and association analysis approach evaluated here continues to be of great practical importance.
Acknowledgments
We gratefully acknowledge support for this research from the National Institutes of Health (NEI R03-EY015216 (to S.S.), NIMH R01-MH595228 (to E.R.H.), NIA R01-AG20135 (to E.R.M.)) and the Neurosciences Education and Research Foundation (support for E.R.H.).
Appendix
Let Z1, Z2 denote the affection status for sibling one and two in a pedigree, with values 1 for affected and 0 for unaffected siblings. Let ζ1, ζ2 denote the sibling covariate vectors, whose components may include X1, X2 as genotypes at the disease susceptibility locus (with alleles A and a and covariate coding according to an assumed inheritance model), Y1, Y2 as continuous covariates, and product terms X1Y1, X2Y2 for GxE interaction. The vectors may include all of these terms or some subset of them, depending on the simulation model.
The sibling recurrence risk ratio λs is defined as follows:
where P(ζ1, ζ2) is the joint probability function of the sibling covariates and the disease probabilities are functions of the β parameters in the logistic regression model equation (1), the disease prevalence K and the frequency p of the susceptibility allele A:
β0 is determined as the solution to the equation
Table A1 shows the joint genotype probabilities for a bi-allelic susceptibility locus for the two siblings.
1. For Model 1 (QTL), let Y1, Y2 denote the continuous covariates for the two siblings. To put boundaries on the simulated covariate values and associated risk increases, we need a mechanism to define the one-unit increase in covariate values to which OR(E) = exp(β2) from equation (1) applies. Let ρ1 denote the proportion proportion of the population at the reference level (baseline risk), with a SIMLA default of 0.0228 [7] that can be modified by the user. Let θ1 denote the corresponding percentile of the mixture normal distribution defined by the three genotype-specific normal distributions and the QTL allele frequency, i.e., P(T ≤ θ1) = ρ1 with T being a random variable following the mixture normal distribution. The SIMLA default is to assume that the same proportion of the population experiences the maximum possible risk increase, i.e. P(T ≥ π) = ρ1. Individuals with originally simulated covariate values y1 ≤ θ1 are assigned the value 0, and individuals with originally simulated covariate values y1 ≥ π are assigned the value ymax, which is determined by the algorithm as P(T ≥ ymax) = ρ1/2. Finally, ρ2 is the proportion of the population with at least a one-unit increase in covariate values, with θ2 denoting the corresponding percentile of the mixture normal distribution, i.e., P(T ≥ θ2) = ρ2. For example, if θ2 is specified as the 80th percentile, 20% of the population have a risk increase of at least exp(β2), compared to the baseline risk. In summary, we have
and analogously for Y2. We assume that sibling phenotypes are conditionally independent, given QTL genotypes and corresponding realized covariate (trait) values. If X1, X2 denote the QTL genotypes for sibling one and two and f denotes the normal distribution density function, the formula for λs is:
2. For Model 2 (GxE interaction), the derivation is similar to Model 1, except that there is only a single normal covariate distribution and the percentiles of interest can be calculated from the standard normal distribution. Thus, the formula for λs is:
In this model, it is possible to incorporate sibling correlations of environmental covariates, in addition to the genotype correlations due to Mendelian inheritance.
3. For Model 3 (heterogeneity), the calculations are simpler since only a main genetic effect is assumed to exist and no integration over the continuous covariate (trait) distribution is necessary. Thus, the formula for λs is:
Table A1.
Joint genotype probabilities for two siblings
| X1 | X2 | P(X1, X2) |
|---|---|---|
| AA | AA | 0.25 p2 (1 + p)2 |
| AA | Aa | 0.5 p2 (1 − p2) |
| AA | aa | 0.25 p2 (1 − p)2 |
| Aa | AA | 0.5 p2 (1 − p2) |
| Aa | Aa | p(1 − p)(1 + p(1 − p)) |
| Aa | aa | p(1 − p)2(1 − 0.5 p) |
| aa | AA | 0.25 p2 (1 − p)2 |
| aa | Aa | p(1 − p)2(1 − 0.5 p) |
| aa | aa | (1 − p)2(1 − 0.5 p)2 |
p is the population frequency of disease susceptibility allele A.
References
- 1.Li M, Boehnke M, Abecasis GR. Efficient study designs for test of genetic association using sibship data and unrelated cases and controls. Am J Hum Genet. 2006;78:778–792. doi: 10.1086/503711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Risch N, Teng J. The relative power of family-based and case-control designs for linkage disequilibrium studies of complex human diseases: DNA pooling. Genome Res. 1998;8:1273–1288. doi: 10.1101/gr.8.12.1273. [DOI] [PubMed] [Google Scholar]
- 3.Teng J, Risch N. The relative power of family-based and case-control designs for linkage disequilibrium studies of complex human diseases. II. Individual genotyping. Genome Res. 1999;9:234–241. [PubMed] [Google Scholar]
- 4.Epstein MP, Veal CD, Trembath RC, Barker JN, Li C, Satten GA. Genetic association analysis using data from triads and unrelated subjects. Am J Hum Genet. 2005;76:592–608. doi: 10.1086/429225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg CR, Umbach DM. A hybrid design for studying genetic influences on risk of diseases with onset early in life. Am J Hum Genet. 2005;77:627–636. doi: 10.1086/496900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt MA, Hauser ER, Martin ER, Schmidt S. Extension of the SIMLA package for generating pedigrees with complex inheritance patterns: Environmental covariates, gene-gene and gene-environment interaction. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1133. Article 15 [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 9.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M. Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol. 2004;27:53–63. doi: 10.1002/gepi.20000. [DOI] [PubMed] [Google Scholar]
- 11.Fingerlin TE, Boehnke M, Abecasis GR. Increasing the power and efficiency of disease-marker case-control association studies through use of allele-sharing information. Am J Hum Genet. 2004;74:432–443. doi: 10.1086/381652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaid DJ. General score tests for associations of genetic markers with disease using cases and their parents. Genet Epidemiol. 1996;13:423–449. doi: 10.1002/(SICI)1098-2272(1996)13:5<423::AID-GEPI1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Sturmer T, Gefeller O, Brenner H. A computer program to estimate power and relative efficiency to assess multiplicative interactions in flexibly matched case-control studies. Comput Methods Programs Biomed. 2004;74:261–265. doi: 10.1016/j.cmpb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Saunders CL, Barrett JH. Flexible matching in case-control studies of gene-environment interactions. Am J Epidemiol. 2004;159:17–22. doi: 10.1093/aje/kwg250. [DOI] [PubMed] [Google Scholar]
- 15.Chung RH, Hauser ER, Martin ER. Interpretation of simultaneous linkage and family-based association tests in genome screens. Genet Epidemiol. 2007;31:134–142. doi: 10.1002/gepi.20196. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt S, Schmidt MA, Qin X, Martin ER, Hauser ER. Linkage analysis with gene-environment interaction: model illustration and performance of ordered subset analysis. Genet Epidemiol. 2006;30:409–422. doi: 10.1002/gepi.20152. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt S, Qin X, Schmidt M, Martin ER, Hauser ER. Interpreting analyses of continuous covariates in affected sibling pair linkage studies. Genet Epidemiol. 2007 April 4; doi: 10.1002/gepi.20227. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Qin X, Schmidt S, Schmidt M, Martin E, Hauser E. International Genetic Epidemiology Society. Tampa, FL: 2006. Nov 16–17, A visualization tool for genetic parameters in complex human traits. [Google Scholar]
- 19.Schmidt S, Hauser MA, Scott WK, Postel EA, Agarwal A, Gallins P, Wong F, Chen YS, Spencer K, Schnetz-Boutaud N, Haines JL, Pericak-Vance MA. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78:852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 22.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANG-PTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007 doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou X, Schmidt S, Hauser ER. Evaluation of GIST and LAMP in the GAW15 simulated data. BMC Genetics. doi: 10.1186/1753-6561-1-s1-s41. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]