Fig. 1.
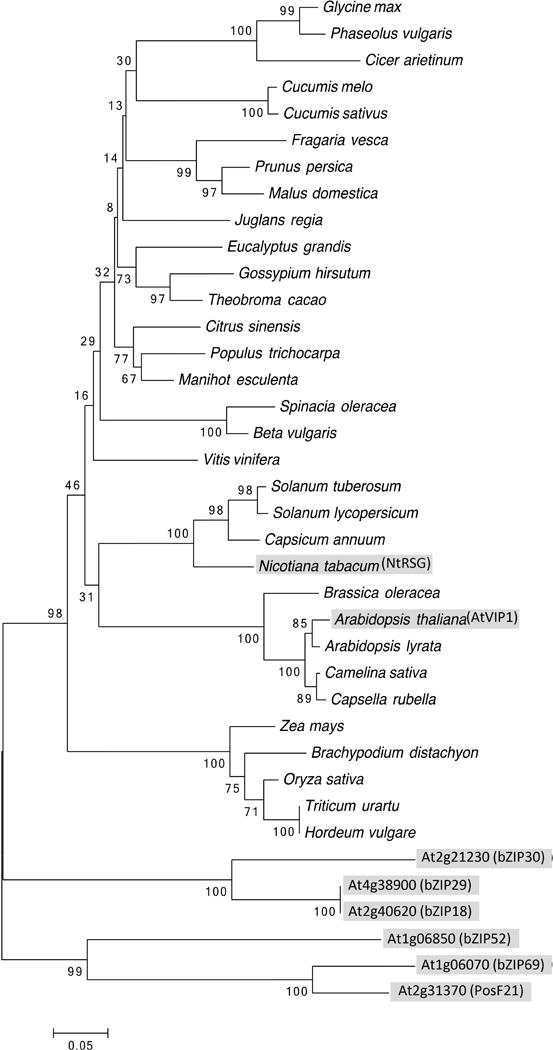
Phylogenetic tree of AtVIP1, its orthologs from representative plant species, and Arabidopsis homologs from the same subgroup. AtVIP1, its 6 closest A. thaliana homologs, and N. tabacum ortholog are highlighted by shaded boxes. The evolutionary history was inferred using the Neighbor-Joining method (Saitou & Nei, 1987). The optimal tree with the sum of branch length = 3.60596289 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl & Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 38 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 179 positions in the final dataset. Evolutionary analyses were conducted using the Molecular Evolutionary Genetics Analysis tool (MEGA, version 6.0.5 for Mac OS) (http://www.megasoftware.net) (Tamura et al., 2013), which also generated this description of the analysis. Scale bar, 0.05 amino acid substitutions per site.
